Abstract
NGN (neurogenin), a proneural bHLH (basic helix–loop–helix) transcription factor, plays a central role in promoting neuronal specification and differentiation in many regions of the central nervous system. NGN activity has been shown extensively to be controlled at the transcriptional level. However, in addition, recent findings have indicated that the levels of NGN protein may also be regulated. In the present study, we have demonstrated that NGN protein stability was regulated in both Xenopus embryos and P19 embryonal carcinoma cells, a mammalian neuronal model system. In both systems, NGN was a highly unstable protein that was polyubiquitinated for destruction by the proteasome. NGN binds to DNA in complex with its heterodimeric E-protein partners E12 or E47. We observed that NGN was stabilized by the presence of E12/E47. Moreover, NGN was phosphorylated, and mutation of a single threonine residue substantially reduced E12-mediated stabilization of NGN. Thus E-protein partner binding and phosphorylation events act together to stabilize NGN, promoting its accumulation when it can be active.
Keywords: neurogenin, polyubiquitination, protein stability, ubiquitin–proteasome system, Xenopus neuronal differentiation
Abbreviations: (b)HLH, (basic) helix–loop–helix; cdk, cyclin-dependent kinase; CHX, cycloheximide; CKII, casein kinase II; GFP, green fluorescent protein; HA, haemagglutinin; HRP, horseradish peroxidase; IVT, in vitro translated; NeuroD, neurogenic differentiation; NGN, neurogenin; IVT 35S-NGN, NGN IVT in the presence of 35S-methionine; NGNR1, NGN-related 1; Ni-NTA, Ni2+-nitrilotriacetate; NP40, Nonidet P40; UPS, ubiquitin–proteasome system; WT, wild-type
INTRODUCTION
Cell cycle exit and differentiation of neurons is a tightly regulated process, involving the activity of an array of transcription factors, including the bHLH (basic helix–loop–helix) family of proneural proteins. One of the key molecules within this family is NGN (neurogenin), which is essential for neurogenesis in the brain, sensory ganglia and spinal cord [1–3]. In Xenopus, NGNR1 [NGN-related 1; hereafter referred to as NGN] is the earliest marker of primary neuron differentiation [4] when the first neurons differentiate out of the neural plate. NGN is most closely homologous with mammalian NGN2, based on sequence and function, with mammalian NGN2 also found to be essential for development of the central nervous system [1].
The regulation of NGN expression and transcriptional activation has been studied extensively [4], but, until recently, regulation of NGN protein levels has not been investigated. We have shown previously that, in Xenopus, the cdk (cyclin-dependent kinase) inhibitor p27Xic1 promotes neurogenesis at least partially by stabilizing NGN [5] and, more recently, p27kip1 has been shown to stabilize mouse NGN2 [6], although interestingly the activity of these cdk inhibitors appears to be independent of their ability to regulate the cell cycle. Thus NGN appears to be an unstable protein, although the mechanism regulating its stability is unknown.
One of the major pathways for degrading cellular proteins is the UPS (ubiquitin–proteasome system; reviewed in [7]). This form of regulated protein destruction requires the covalent attachment of the small protein ubiquitin to amino groups of the targeted substrates, usually on to lysine residues. Subsequent ubiquitin molecules are added to the first via lysine residues within ubiquitin itself to form a polyubiquitin chain. Polyubiquitinated proteins are then transported to the 26 S proteasome and are degraded. A growing number of factors promoting cell proliferation or cell differentiation have been shown to be targeted by the UPS. In particular, the bHLH factor MyoD (myogenic differentiation), a determinant of vertebrate muscle differentiation, is regulated by the UPS [8], as are the proliferative HLH proteins Id1, Id2 and Id3 [9]. However, evidence for the ubiquitin-mediated proteolysis of proneural proteins is limited [10], and a role for ubiquitin-mediated proteolysis in the control of NGN stability has not been investigated.
In the present study, using extracts from Xenopus eggs and embryos and the P19 cell line that is competent to differentiate into neurons, we have demonstrated that the stability of NGN was regulated by ubiquitin-mediated proteolysis. NGN had a very short half-life in eggs and embryos, and proliferating P19 cells. It was stabilized by inhibitors of the proteasome and by inhibitors of polyubiquitination. Moreover, polyubiquitinated forms of NGN were readily detected. NGN is thought to be transcriptionally active only when bound to its heterodimeric binding partner E-protein (E12/E47). We have shown that the addition of E12/E47 significantly increased the half-life of NGN in both Xenopus extracts and P19 cells. Previously, E47 has been shown to regulate phosphorylation of the related bHLH proneural factor Mash1 on a specific CKII (casein kinase II)-target site, which also regulates Mash1 stability [11]. A similar CKII site was present in NGN, and we have shown that, when it was mutated, E12-mediated stabilization of NGN was severely impeded, indicating that the phosphorylation state also regulated NGN levels. Thus NGN protein has a very short half-life, but this can be significantly lengthened by E12/E47, demonstrating a mechanism allowing NGN protein to accumulate specifically when it is competent to activate transcription.
MATERIALS AND METHODS
Plasmids
NGN [4] and Xenopus NeuroD (neurogenic differentiation) [12] were cloned into pCS2. NGN T118A mutant was generated by site-directed mutagenesis using the QuikChange® multi-site-directed mutagenesis kit (Stratagene), with NGN cloned into pCS2 as the template and 5′-GAAGATGCCAAACTCGCCAAGATAGAGACCTTGCGC-3′ as the primer. For bacterial expression, NGN was cloned into pET-30a. For mammalian studies, a HA (haemagglutinin) tag was added to NGN by PCR and cloned into pCS2 using the primers 5′-CGCGGATCCACCATGTTCGTCAAATCTGAGACTCTGG-3′ and 5′-GCGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTAGATACAGTCCCTGGCGAGG-3′. Xenopus E12, mouse E47 and FLAG–ubiquitin constructs have been described previously [13–15].
Preparation of Xenopus egg and embryo extracts
Xenopus laevis eggs and neurula stage embryos were obtained by standard methods and staged according to Nieuwkoop and Faber [16]. Extracts were prepared as described previously [17], with minor modifications. De-jellied eggs or embryos were washed in chilled XB buffer [100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes (pH 7.7) and 50 mM sucrose] and mixed with 10 mg/μl cytochalasin B (Sigma). Eggs and embryos were packed by centrifugation for 1 min at 4 °C at 100 g and 400 g respectively, and then ruptured by centrifugation (5 min at 16000 g at 4 °C). Out of the three phases formed, the intermediate cytoplasmic layer was removed and spun twice (5 min at 16000 g at 4 °C). The extracts were supplemented with LPC (leupeptin, pepstatin and chymostatin), each at 10 μg/ml, and a one-twentieth volume of energy mixture (150 mM creatine phosphate, 20 mM ATP, 2 mM EGTA and 20 mM MgCl2) and were used immediately.
In vitro transcription and translation
NGN, NGN T118A and NeuroD were IVT (in vitro transcribed and translated) using 35S-methionine (GE Healthcare) and the TnT® SP6 Quick Coupled transcription/translation system (Promega). GFP (green fluorescent protein) and E12 protein were obtained similarly using non-radiolabelled methionine.
In vitro degradation assays
Degradation assays were performed as described previously [18], with minor modifications. Extracts were supplemented with 100 μg/ml CHX (cycloheximide; Sigma) and 1.25 mg/ml ubiquitin (Sigma) or 10 mg/ml methylated ubiquitin (BioMol). When indicated, 200 μM MG132 (BioMol) was added for 5 min to the extracts before addition of NGN or NeuroD. 35S-Labelled proteins were mixed with extracts and incubated at 20 °C. For degradation assays in the presence of E12, unlabelled IVT E12 or GFP was added where indicated. Aliquots were taken at various time points and mixed with Laemmli sample buffer. Samples were denatured for 3 min at 95 °C and separated by SDS/PAGE. Gels were dried and proteins were visualized by phosphorimaging and autoradiography.
In vitro ubiquitination assay
Ubiquitination assays were conducted as described previously [19], with minor modifications. IVT 35S-NGN (NGN IVT in the presence of 35S-methionine) was mixed with interphase egg or neurula stage embryo extracts supplemented with 200 μM MG132 and either 2.5 mg/ml His–ubiquitin (His-tagged ubiquitin) or 1.25 mg/ml ubiquitin, and incubated at room temperature (20 °C) for 1 h. The reaction was then diluted 10-fold in His buffer [100 mM Tris/HCl (pH 7.4), 1% (v/v) NP40 (Nonidet P40), 8 M urea, 20 mM imidazole, 600 mM NaCl and 10% (v/v) ethanol] and Complete protease inhibitor cocktail (Roche) and 1 μM pepstatin were added before mixing with Ni-NTA (Ni2+-nitrilotriacetate)–agarose beads (Qiagen). After incubation for 90 min at room temperature, beads were washed several times with His buffer, and proteins were eluted with Laemmli sample buffer. Samples were analysed as described above.
Ubiquitination assay of bacterially expressed NGN
NGN was cloned into the pET-30a vector to allow the expression of a C-terminus His6-tagged protein (NGN–His) in Escherichia coli. NGN–His was expressed in Rosetta cells and purified using the Ni-NTA–agarose resin (Qiagen), according to the manufacturer's instructions. Since NGN–His was expressed in inclusion bodies, purification was performed in the presence of urea. Briefly, proteins were solubilized in N300 buffer [50 mM Tris/HCl (pH 9.4), 500 mM NaCl, 1 mM PMSF, 10 mM 2-mercaptoethanol] supplemented with 8 M urea. Ni-NTA–agarose beads were added to the protein and incubated for 1 h at room temperature. Beads were washed successively with N300 buffer containing 4 M urea, N300 buffer containing 1 M urea and, finally, transferred into XB buffer. NGN–His still bound to the beads was then incubated with Xenopus activated egg extract together with ubiquitin (1.25 mg/ml) and MG132 (700 μM) for 1 h at 20 °C. NGN–His was then re-purified by first diluting the reactions 10-fold in a Urea buffer [100 mM Tris/HCl (pH 7.4), 1% Igepal CA-360 (Sigma), 8 M urea, 20 mM imidazole and 600 mM NaCl] and incubating at room temperature for 2 h. Beads were washed several times with Urea buffer, and proteins were eluted with Laemmli sample buffer. Samples were analysed by Western blotting. Ubiquitinated NGN was detected using an anti-(mono- and poly-ubiquitinated conjugates) antibody (FK2 clone; BioMol) and an HRP (horseradish peroxidase)-conjugated anti-(mouse IgG) antibody (GE Healthcare).
Cell culture and transfections
P19 cells were grown as described previously [20]. Transfections were performed on cells grown on poly-L-lysine (Sigma)-coated plates with NGN–HA, pCS2 and/or E47 constructs using Lipofectamine2000™ (Invitrogen). Degradation assays were performed 24 h after transfection. Protein synthesis was blocked with 10 μM CHX (t=0) and samples were collected at intervals. Where indicated, 20 μM MG132 was used. Cells were lysed in lysis buffer [50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1% (v/v) NP40, Complete protease inhibitor cocktail and 1 μM pepstatin] and subjected to Western blot analysis using 20 μg of total proteins. NGN protein was detected using an anti-HA antibody (Roche) and HRP-conjugated anti-(mouse IgG) antibody (GE Healthcare).
For immunoprecipitation assays, NGN–HA or untagged NGN was co-transfected with FLAG–ubiquitin in P19 cells. At 24 h post-transfection, cells were incubated in the presence of 50 μM MG132 for 1 h, and cell extracts were prepared in lysis buffer. NGN was immunoprecipitated in IP buffer [50mM Tris/HCl (pH 7.5), 150 mM NaCl, 0.1% NP40, complete protease inhibitor cocktail and 1 μM pepstatin] using a HA–matrix (Roche) and purified by competition with the HA–peptide (Roche). Ubiquitinated proteins were immunoprecipitated using an anti-FLAG antibody (M2 clone; Sigma) and Protein A–Sepharose (GE Healthcare). Purified proteins were subjected to Western blot analysis using an HRP-coupled anti-HA antibody (Roche) or HRP-coupled anti-FLAG antibody (M2 clone; Sigma).
Phosphatase treatment
Lambda phosphatase (New England Biolabs) was used according to the manufacturer's instructions. IVT 35S-NGN was incubated in the presence of E12 and egg extract for 1 h, and 200 μM MG132 was added before treatment with lambda phosphatase. As a control, buffer alone with no enzyme was used. Samples were analysed as described above. For P19 cells, NGN–HA and E47 were transfected into cells and, 24 h later, protein synthesis was blocked and samples were collected after 5 min. Western blot analysis was performed as described above.
RESULTS
NGN protein is highly unstable
NGNs, particularly Xenopus NGNR1 and mammalian NGN2, are crucially important proteins in the development of the nervous system. However, despite this, very little is known about the regulation of NGN at the protein level. In recent years, two studies have indicated that NGN is unstable [5,6]; however, neither the half-life of the protein nor the mechanism by which it is turned over has been investigated in detail.
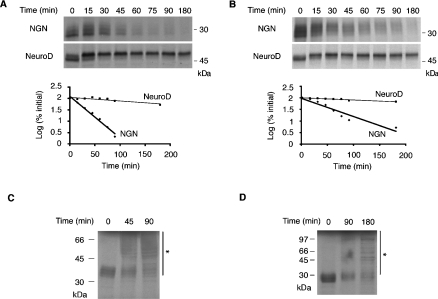
Extracts from Xenopus eggs and embryos have been widely used to study proteolysis of several proteins [19]. We have demonstrated previously that NGN protein stability is regulated in developing Xenopus embryos [5], so we have initially taken an extract approach to investigate NGN degradation biochemically. IVT 35S-NGN was incubated in extracts prepared from activated interphase Xenopus eggs or neurula stage embryos, aliquots were removed at intervals and the amount of NGN protein remaining was determined by SDS/PAGE, autoradiography and quantitative phosphorimaging analysis. NGN was highly unstable in interphase egg and embryo extracts, with a half-life of 21.9±2.2 and 37.9±4.0 min (values are means±S.E.M.) respectively (Figures 1A and 1B). Although the half-life of NGN in embryos was almost twice as long as in eggs, it appeared likely that the mechanism of proteolysis was the same in both systems (see below). We wished to determine whether other proneural proteins were unstable in Xenopus eggs and embryos or if NGN protein was degraded specifically. To this end, we measured the stability of the closely related proneural protein NeuroD, itself a transcriptional target of NGN [12]. Strikingly, in both egg and neurula stage embryo extracts, NeuroD was remarkably stable, with half-lives of 190.7±22.9 and 262.3±11.4 min respectively (Figures 1A and 1B). From these results, it is clear that, although NGN is rapidly degraded in both eggs and embryos, NeuroD is significantly more stable.
Figure 1. NGN is a highly unstable protein.
(A, B) Extracts were prepared from interphase eggs (A) or neurula stage embryos (B) and supplemented with IVT 35S-NGN or 35S-NeuroD. Samples were taken at the time points indicated and subjected to SDS/PAGE. Gels were analysed by autoradiography (top and middle panels) and quantitative phosphorimaging analysis (bottom panels). (C, D) Long exposure of IVT 35S-NGN in egg (C) and embryo (D) extracts separated by SDS/PAGE to reveal higher-molecular-mass forms of NGN (asterisks). Molecular-mass markers (in kDa) are indicated.
Covalent attachment of polyubiquitin chains is frequently used to target proteins for degradation. To detect such forms, we incubated IVT 35S-NGN in egg and embryo extracts for increasing time periods and performed a long autoradiographic exposure of the gel. A ladder of higher-order forms of NGN, stretching up to the top of the gel, was observed that may correspond to polyubiquitinated forms of the protein (asterisks in Figures 1C and 1D).
NGN is degraded by ubiquitin-mediated proteolysis
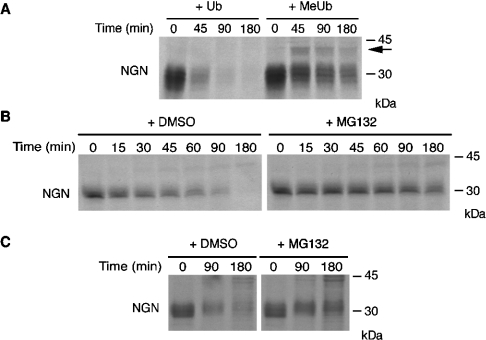
If NGN destruction requires polyubiquitination, it should be stabilized by the addition of methylated ubiquitin, a chemically modified form of ubiquitin that cannot form polyubiquitin chains [21]. Methylated ubiquitin was added to egg extracts in the presence of IVT 35S-NGN, and aliquots were removed at intervals for electrophoretic analysis. Addition of methylated ubiquitin resulted in substantial stabilization of NGN protein even up to 180 min (Figure 2A). Moreover, a prominent mono-ubiquitinated form, running approx. 10 kDa slower by SDS/PAGE, was clearly visible (arrow in Figure 2A), again demonstrating that NGN protein was directly ubiquitinated.
Figure 2. NGN is degraded by the UPS.
(A) A degradation assay of IVT 35S-NGN was performed with interphase egg extracts in the presence of ubiquitin (Ub) or methylated ubiquitin (MeUb), and was analysed by SDS/PAGE and autoradiography. The arrow indicates mono-ubiquitinated NGN. (B, C) Extracts were prepared from interphase eggs (B) or neurula stage embryos (C) and supplemented with IVT 35S-NGN in the presence of the proteasome inhibitor MG132 (200 μM) or DMSO alone. Samples were taken at the time points indicated and were subjected to SDS/PAGE and subsequent autoradiography. Molecular-mass markers (in kDa) are indicated on the right.
Polyubiquitination is frequently used as a signal to direct proteins to the proteasome for degradation [22]. To investigate whether NGN is subject to proteasomal degradation, we performed degradation assays of IVT 35S-NGN in the presence of a chemical inhibitor of the proteasome, MG132. When MG132 was added to egg extracts, NGN was stabilized significantly, with the half-life increasing from 21.9±2.2 to 124.0±19.1 min (Figure 2B). Moreover, a similar stabilizing effect was seen in extracts from neurula stage embryos, extending the half-life from 37.9±4.0 to 123.8±14.6 min (Figure 2C), demonstrating that the mechanism of destruction is the same at these different developmental stages. A similar stabilization was also seen with other proteasome inhibitors, including MG262, vinyl sulfone and epoxomycin (results not shown). Therefore polyubiquitinated NGN is degraded by the proteasome.
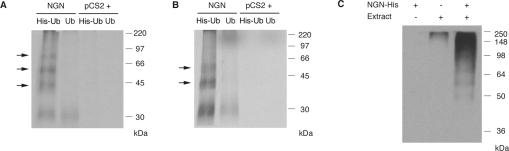
To demonstrate conclusively that NGN is polyubiquitinated, we set out to isolate ubiquitin-bound forms of the NGN protein. To this end, we incubated IVT 35S-NGN in egg extracts in the presence of MG132 and His–ubiquitin. Proteins covalently modified with His–ubiquitin were subsequently isolated using Ni2+-NTA–agarose beads. To ascertain that only proteins covalently bound to ubiquitin would be pulled down and since NGN is known to bind proteins that can themselves be polyubiquitinated [9,23], a buffer containing 8 M urea was used to wash the beads before elution. Purified proteins were then separated by SDS/PAGE for autoradiographic analysis. NGN was covalently associated with His–ubiquitin, and slower migrating forms that correspond to polyubiquitinated species were clearly visible (Figure 3A). Similar polyubiquitinated forms of NGN were generated when NGN was incubated with His–ubiquitin in neurula stage embryo extracts (Figure 3B). In contrast, only very low background levels of associated protein was observed when ubiquitin without the His tag was added. Similarly, no radiolabelled proteins were purified when an IVT reaction containing the vector alone was added to the extract in place of IVT 35S-NGN (Figures 3A and 3B).
Figure 3. NGN is polyubiquitinated.
(A, B) IVT 35S-NGN was incubated with interphase egg (A) or neurula stage embryo (B) extracts in the presence of ubiquitin (Ub) or His–ubiquitin (His–Ub), and ubiquitin-bound proteins were purified by affinity chromatography with Ni2+-NTA–agarose. The empty vector (pCS2+) was used as control. The arrows indicate polyubiquitinated forms of NGN. (C) Bacterially expressed and purified NGN–His was incubated with interphase egg extracts in the presence of ubiquitin and MG132 and re-isolated before elution. Western blotting was performed with an anti-ubiquitin antibody. Molecular-mass markers (in kDa) are indicated on the right.
Another approach was taken to show directly that NGN is bound to ubiquitin. NGN–His protein was expressed in bacteria, purified with Ni2+-NTA–agarose and incubated, while still bound to the beads, with egg extract in the presence of MG132. NGN–His was then re-isolated and washed in buffer containing 8 M urea before elution. Proteins were then separated by SDS/PAGE, and Western blot analysis was performed using an antibody to detect ubiquitin. Polyubiquitin chains of different lengths were specifically detected after NGN–His had been passed through the egg extract (Figure 3C). Thus NGN is clearly polyubiquitinated and degraded by the proteasome in Xenopus eggs and embryos.
NGN is stabilized by its heterodimeric binding partner E12
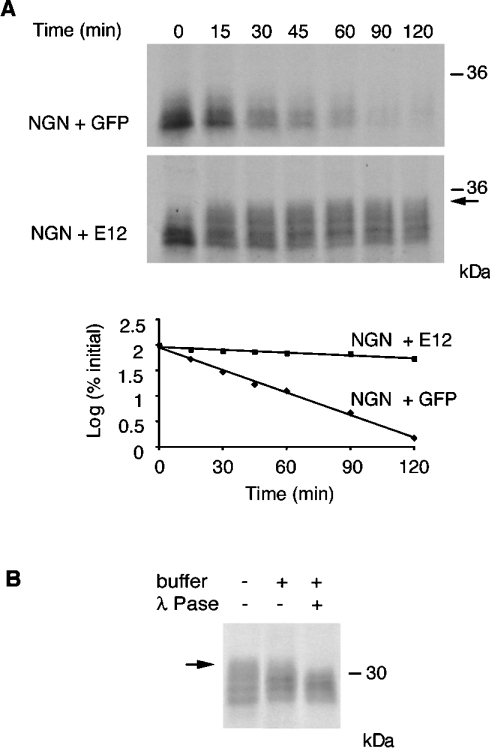
NGN is a highly unstable protein, subject to very rapid ubiquitin-mediated proteolysis, yet its transcriptional activity is vital to promote neurogenesis and repress gliogenesis in a wide number of regions in the mammalian central nervous system [3]. To act as a transcription factor, NGN binds to DNA as a heterodimer with the ubiquitous splice products of the bHLH E2A gene E12 and E47 [1,3]. It has been shown previously that, in some circumstances, the related bHLH proneural factor Mash1 can be stabilized by the presence of E47 [11]. We thus determined whether NGN was similarly stabilized by E12/E47. To this end, we incubated IVT 35S-NGN, in the absence or presence of IVT-unlabelled E12, in egg extracts and removed aliquots at intervals for autoradiographic analysis. Co-incubation of NGN and E12 in egg extracts led to a substantial stabilization of NGN protein, with the half-life increasing from 21.9±2.2 to 166.4±19.9 min (Figure 4A). In contrast, co-incubation of NGN with GFP as control had no effect.
Figure 4. E12 stabilizes NGN and promotes its phosphorylation.
(A) E12 protein was added to Xenopus interphase egg extracts and the rate of degradation of IVT 35S-NGN was assayed relative to a control supplemented with GFP. Gels were analysed by autoradiography (top and middle panels) and quantitative phosphorimaging analysis (bottom panel). (B) Lambda phosphatase (λ Pase) treatment of NGN incubated with E12. Arrows in (A) and (B) indicate a slower migrating form of NGN that accumulates in the presence of E12. Molecular-mass markers (in kDa) are indicated on the right.
Interestingly, NGN that accumulated in the presence of E12 was significantly retarded in its electrophoretic migration compared with the protein incubated in the presence of GFP (arrow in Figure 4A). It has been shown that the destruction of some transcription factors by ubiquitin-mediated proteolysis, including those of other bHLH factors, such as MyoD, Myf5 and Mash1, is regulated by phosphorylation [11,24,25]. The mobility shift of NGN could thus be consistent with phosphorylation of a stable form of the protein. To confirm that this was the case, we treated extracts containing E12 and NGN with lambda phosphatase. An increase in mobility of the higher-order forms of NGN seen in the presence of E12 was observed under these conditions, resulting in the loss of the upper band (Figure 4B), indicating that E12 does promote accumulation of hyperphosphorylated forms of NGN.
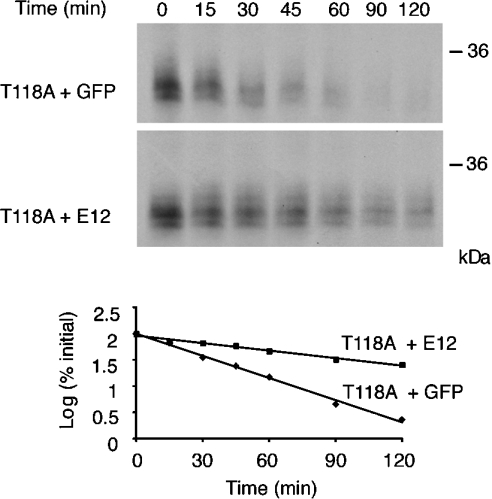
In the case of Mash1, a proneural factor related to NGN, it has been determined that the ubiquitous E-protein E47 promotes CKII-dependent phosphorylation on Ser152 of Mash1 and that this modification increases Mash1 protein stability [11]. We performed a sequence alignment between Mash1 and NGN and identified a similar motif in NGN located in the loop domain. In order to investigate the role of this site in modulating NGN stability, we used site-directed mutagenesis to replace the potentially phosphorylated threonine residue (Thr118) with an alanine residue. Degradation assays were then performed using this mutant in the absence and presence of E12. Strikingly, we found that E12 promoted the accumulation of WT (wild-type) NGN, but was less effective at inhibiting the degradation of T118A NGN, whereas both proteins displayed similar stability in the absence of E12 (Figure 5). The half-lives in the absence of E12 were similar (21.9±2.2 and 23.3±2.6 min for WT and T118A NGN respectively), but differed significantly in the presence of E12 (61.5±5.3 min for T118A compared with 166.4±19.9 min for WT). Interestingly, the slowest migrating form of NGN seen when WT NGN and E12 were co-incubated did not accumulate significantly when using the T118 mutant (compare Figure 4A, arrow, with Figure 5), indicating a differing phosphorylation status of the two proteins, hyperphosphorylation of NGN being potentially associated within enhanced stability.
Figure 5. Reduced stability of a CKII mutant of NGN in the presence of E12.
E12 protein was added to Xenopus interphase egg extracts and the rate of degradation of IVT 35S-NGN T118A was assayed relative to a control supplemented with GFP. Gels were analysed by autoradiography (top and middle panels) and quantitative phosphorimaging analysis (bottom panel). Molecular-mass markers (in kDa) are indicated on the right.
NGN is unstable in P19 cells
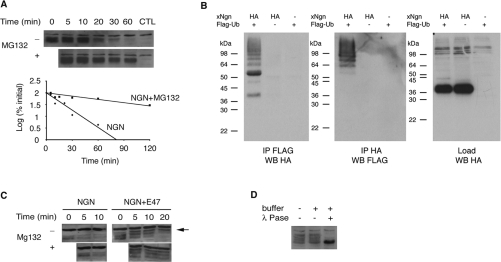
The experiments described above demonstrate that NGN is a highly unstable protein in both Xenopus eggs and embryos. To assess whether the same is true in mammalian cells, we investigated NGN stability in P19 embryonal carcinoma cells, a cell line that is competent to differentiate into neurons under the influence of bHLH transcription factors [26]. A plasmid expressing HA-tagged NGN was transfected into P19 cells and, at t=0, CHX was added to stop ongoing protein synthesis. Samples were collected at intervals and levels of NGN protein were monitored by Western blotting. Similar to our results in Xenopus, NGN was extremely unstable in P19 cells, having a half-life of just 17.1±1.2 min (Figure 6A). Moreover, this degradation was mediated by the proteasome, as it was effectively blocked by the addition of MG132. The short half-life exhibited by NGN might indicate why it is relatively poor at inducing neural differentiation of P19 cells relative to other bHLH proneural genes tested [26].
Figure 6. NGN stability in P19 cells.
(A) HA–NGN was transfected in P19 cells and, after protein synthesis was inhibited with CHX, samples were collected at intervals and subjected to Western blotting with an anti-HA antibody (top and middle panels). Control (CTL) corresponds to cells transfected with untagged NGN. The assay was performed in the presence or absence of 20 μM MG132 to block proteasome activity. Quantification was performed by densitometry scanning (lower panel). (B) HA–NGN was co-transfected with FLAG–ubiquitin and reciprocal immunoprecipitations were carried out. Ubiquitinated proteins were immunoprecipitated (IP) with an anti-FLAG antibody and subjected to Western blotting (WB) with an anti-HA antibody (left-hand panel). HA-tagged proteins were pulled down using an HA–matrix and subjected to Western blotting with anti-FLAG antibody (middle panel). One-tenth of the loading material (Load) was also subjected to Western blotting (right-hand panel). Controls used were P19 cells transfected with HA–NGN in the absence of FLAG–ubiquitin and untagged NGN in the presence of FLAG–ubiquitin. Molecular-mass markers (in kDa) are indicated on the left. (C) Stabilization of NGN by E47. P19 cells were transfected with HA–NGN in the presence or absence of E47, and a degradation assay was performed as described in (A). The arrow indicates a slower migrating form of NGN only seen in the presence of E47. (D) Lambda phosphatase (λ Pase) treatment of NGN transfected into the cells with E47.
To demonstrate that NGN is ubiquitinated in P19 cells, HA–NGN was co-transfected with FLAG–ubiquitin. Ubiquitinated proteins were then immunoprecipitated with an anti-FLAG antibody and subjected to Western blotting with an anti-HA antibody. A protein ladder was clearly visible, indicating that HA–NGN was polyubiquitinated (Figure 6B). Similarly, when HA–NGN was pulled down using an HA–matrix and subjected to Western blotting with an anti-FLAG antibody to detect ubiquitin, a protein ladder of high-molecular mass was observed (Figure 6B). This demonstrates clearly that NGN is polyubiquitinated in P19 cells.
Co-incubation with E12 in Xenopus egg extracts leads to accumulation of hyperphosphorylated forms of NGN. To determine if a similar stabilization occurs in mammalian cells, P19 cells were co-transfected with NGN and the splice isoform E47. The presence of E47 also resulted in a significant lengthening of the half-life of the NGN protein in P19 cells (Figure 6C). Again, distinct slower migrating forms of NGN accumulated, including a more slowly migrating form of NGN seen in the presence of E47, which was not seen in its absence (Figure 6C, arrow). Treatment with lambda phosphatase demonstrated that these slower migrating forms of NGN were indeed hyperphosphorylated (Figure 6D).
Therefore NGN protein is regulated similarly in Xenopus eggs, embryos and P19 cells, where it is degraded by the UPS. Degradation is inhibited by co-expression of E12/E47 and this results in accumulation of hyperphosphorylated forms of the protein.
DISCUSSION
Over the past decade, it has become increasing clear that a broad range of regulatory proteins, including transcription factors, are controlled at the level of protein stability by the UPS [22,27]. Neural determination and differentiation are controlled by proneural proteins. Although numerous studies have investigated their regulation at the gene expression level, very little is known about regulation of the proneural proteins themselves. In the present study, we have demonstrated that one of the most prominent proneural proteins, NGN, is regulated at the level of protein stability.
NGN is a member of the bHLH class of transcription factors. Within this class, the most studied is the bHLH protein MyoD, which plays a pivotal role in regulating myogenesis. MyoD is degraded by the UPS [8], and is subject to more than one mode of ubiquitination [28], probably by several E3 ligases. However, pathways controlling its destruction are far from fully understood. Moreover, the myogenic bHLH Myf5 is also degraded by the UPS, but its regulation probably differs significantly from MyoD [24]. It is becoming clear that we will understand the regulation of destruction of each protein only by studying them on a case-by-case basis. Previously, only one proneural protein, Mash1, has been shown to be degraded following polyubiquitination [11] and, as with myogenic factors, the limited amount of data available makes it hard to draw general conclusions. In the present study, we show that the NGN protein has a short half-life, is polyubiquitinated and degraded by the proteasome in Xenopus eggs and embryos, and in mammalian P19 embryonal carcinoma cells. Interestingly, we show that a related bHLH protein, NeuroD, is stable in Xenopus eggs and embryos.
During development, NGN expression is transient, and prolonged up-regulation by mRNA microinjection can result in apoptosis [29]. NeuroD is downstream of NGN and drives terminal differentiation of neurons [12]. Hence it may be more desirable for the early factor NGN to have a short half-life, whereas the factor driving cell cycle exit and terminal differentiation, NeuroD, is more long-lived.
Polyubiquitination of many proteins targeted for destruction by the UPS is regulated by phosphorylation. This is certainly true of members of the bHLH family, such as MyoD, Myf5 and E12 [8,23,24,30]. MyoD and Myf5 stability is controlled by phosphorylation mediated by cdks and also appears to be cell-cycle-regulated [24,25,31]. We observed that NGN is phosphorylated in both Xenopus and mammalian cells. Co-expression of NGN with E12/E47 lead to NGN stabilization and resulted in accumulation of hyperphosphorylated forms of NGN, in both Xenopus eggs and P19 cells. The stability of the related proneural gene Mash1 is controlled by phosphorylation on a specific CKII site, an event that is itself promoted by binding to an E protein partner [11]. We identified a similar potential CKII site in NGN, namely Thr118. We found that a T118A mutant of NGN had a similar half-life to the WT protein on its own; however, the T118A mutant could no longer be stabilized by co-incubation with E12 and the usual associated hyperphosphorylated forms of NGN were substantially decreased. This indicates that, as in the case of Mash1, E12 may promote phosphorylation on this residue, resulting in increased stabilization of the complex. NeuroD is also phosphorylated in egg extracts, but in this case is almost fully stable, and the functional significance of this phosphorylation is unclear.
The NGN family of transcription factors is crucially important for neurogenesis. Indeed, levels of NGN protein have been experimentally manipulated in model systems with potential therapeutic implications. For instance, recovery from spinal cord injury in a rat model is significantly enhanced when grafted neural precursors have been supplied with ectopic NGN2 [32]. In the present study, we demonstrate for the first time that Xenopus NGN has a strikingly short half-life, both in Xenopus eggs and embryos, and in mammalian P19 cells that are capable of differentiating into neurons. In addition, the half-life of NGN is modulated by its heterodimeric binding partner E12/E47 and by phosphorylation. A detailed understanding both of the potentially multiple mechanisms that regulate NGN stability and identification of the molecules which can target NGN for ubiquitin-mediated proteolysis may not only be important for a thorough understanding of differentiation, but may also have important therapeutic implications if we are to potentiate mechanisms to promote neurogenesis formation in vitro and in vivo. The present study is the first step towards this goal.
Acknowledgments
We thank Professor Kohei Miyazono (Department of Molecular Pathology, Graduate School of Medicine, University of Tokyo, Bunkyo-ku, Tokyo, Japan) for the FLAG–ubiquitin construct. We also thank François Guillemot, Olivier Raineteau, Laurent Nguyen and Shin-ichi Ohnuma for helpful discussions. This work was supported by grants from the U.K. Medical Research Council (G050010) and the Biotechnology and Biological Sciences Research Council (BB/C004108/1).
References
- 1.Bertrand N., Castro D. S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 2.Furlong R. F., Graham A. Vertebrate neurogenin evolution: long-term maintenance of redundant duplicates. Dev. Genes Evol. 2005;215:639–644. doi: 10.1007/s00427-005-0023-x. [DOI] [PubMed] [Google Scholar]
- 3.Ross S. E., Greenberg M. E., Stiles C. D. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 4.Ma Q., Kintner C., Anderson D. J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 5.Vernon A. E., Devine C., Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen L., Besson A., Heng J. I., Schuurmans C., Teboul L., Parras C., Philpott A., Roberts J. M., Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandi D., Tahiliani P., Kumar A., Chandu D. The ubiquitin–proteasome system. J. Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 8.Song A., Wang Q., Goebl M. G., Harrington M. A. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bounpheng M. A., Dimas J. J., Dodds S. G., Christy B. A. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- 10.Shou J., Rim P. C., Calof A. L. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat. Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 11.Vinals F., Reiriz J., Ambrosio S., Bartrons R., Rosa J. L., Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 2004;23:3527–3537. doi: 10.1038/sj.emboj.7600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J. E., Hollenberg S. M., Snider L., Turner D. L., Lipnick N., Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 13.Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., Miyazawa K. Negative regulation of transforming growth factor-β (TGF-β) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23:6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 14.Rashbass J., Taylor M. V., Gurdon J. B. The DNA-binding protein E12 co-operates with XMyoD in the activation of muscle-specific gene expression in Xenopus embryos. EMBO J. 1992;11:2981–2990. doi: 10.1002/j.1460-2075.1992.tb05368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X. H., Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 16.Nieuwkoop P. D., Faber J. Amsterdam: North Holland Publishing; 1967. Normal Table of Xenopus laevis (Daudin) [Google Scholar]
- 17.Murray A. W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 18.Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 19.Salic A., Lee E., Mayer L., Kirschner M. W. Control of β-catenin stability: reconstitution of the cytoplasmic steps of the wnt pathway in Xenopus egg extracts. Mol. Cell. 2000;5:523–532. doi: 10.1016/s1097-2765(00)80446-3. [DOI] [PubMed] [Google Scholar]
- 20.Rudnicki M. A., McBurney M. W. Cell culture methods and induction of differentiation of embryocarcinoma cells. In: Robertson E. J., editor. Teratocarcinomas and Embryonic Stem Cells. Oxford: IRL Press; 1987. pp. 19–49. [Google Scholar]
- 21.Hershko A., Ganoth D., Pehrson J., Palazzo R. E., Cohen L. H. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J. Biol. Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- 22.Ciechanover A., Orian A., Schwartz A. L. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Kho C. J., Huggins G. S., Endege W. O., Hsieh C. M., Lee M. E., Haber E. Degradation of E2A proteins through a ubiquitin-conjugating enzyme, UbcE2A. J. Biol. Chem. 1997;272:3845–3851. doi: 10.1074/jbc.272.6.3845. [DOI] [PubMed] [Google Scholar]
- 24.Doucet C., Gutierrez G. J., Lindon C., Lorca T., Lledo G., Pinset C., Coux O. Multiple phosphorylation events control mitotic degradation of the muscle transcription factor Myf5. BMC Biochem. 2005;6:27. doi: 10.1186/1471-2091-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tintignac L. A., Leibovitch M. P., Kitzmann M., Fernandez A., Ducommun B., Meijer L., Leibovitch S. A. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp. Cell Res. 2000;259:300–307. doi: 10.1006/excr.2000.4973. [DOI] [PubMed] [Google Scholar]
- 26.Farah M. H., Olson J. M., Sucic H. B., Hume R. I., Tapscott S. J., Turner D. L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 27.Collins G. A., Tansey W. P. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo W., Gautier J. XNGNR1-dependent neurogenesis mediates early neural cell death. Mech. Dev. 2005;122:635–644. doi: 10.1016/j.mod.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Nie L., Xu M., Vladimirova A., Sun X. H. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindon C., Albagli O., Domeyne P., Montarras D., Pinset C. Constitutive instability of muscle regulatory factor Myf5 is distinct from its mitosis-specific disappearance, which requires a D-box-like motif overlapping the basic domain. Mol. Cell. Biol. 2000;20:8923–8932. doi: 10.1128/mcb.20.23.8923-8932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofstetter C. P., Holmstrom N. A., Lilja J. A., Schweinhardt P., Hao J., Spenger C., Wiesenfeld-Hallin Z., Kurpad S. N., Frisen J., Olson L. Allodynia limits the usefulness of intraspinal neural stem cell grafts; directed differentiation improves outcome. Nat. Neurosci. 2005;8:346–353. doi: 10.1038/nn1405. [DOI] [PubMed] [Google Scholar]