Abstract
AS160 (Akt substrate of 160 kDa) mediates insulin-stimulated GLUT4 (glucose transporter 4) translocation, but is widely expressed in insulin-insensitive tissues lacking GLUT4. Having isolated AS160 by 14-3-3-affinity chromatography, we found that binding of AS160 to 14-3-3 isoforms in HEK (human embryonic kidney)-293 cells was induced by IGF-1 (insulin-like growth factor-1), EGF (epidermal growth factor), PMA and, to a lesser extent, AICAR (5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside). AS160-14-3-3 interactions were stabilized by chemical cross-linking and abolished by dephosphorylation. Eight residues on AS160 (Ser318, Ser341, Thr568, Ser570, Ser588, Thr642, Ser666 and Ser751) were differentially phosphorylated in response to IGF-1, EGF, PMA and AICAR. The binding of 14-3-3 proteins to HA–AS160 (where HA is haemagglutinin) was markedly decreased by mutation of Thr642 and abolished in a Thr642Ala/Ser341Ala double mutant. The AGC (protein kinase A/protein kinase G/protein kinase C-family) kinases RSK1 (p90 ribosomal S6 kinase 1), SGK1 (serum- and glucocorticoid-induced protein kinase 1) and PKB (protein kinase B) displayed distinct signatures of AS160 phosphorylation in vitro: all three kinases phosphorylated Ser318, Ser588 and Thr642; RSK1 also phosphorylated Ser341, Ser751 and to a lesser extent Thr568; and SGK1 phosphorylated Thr568 and Ser751. AMPK (AMP-activated protein kinase) preferentially phosphorylated Ser588, with less phosphorylation of other sites. In cells, the IGF-1-stimulated phosphorylations, and certain EGF-stimulated phosphorylations, were inhibited by PI3K (phosphoinositide 3-kinase) inhibitors, whereas the RSK inhibitor BI-D1870 inhibited the PMA-induced phosphorylations. The expression of LKB1 in HeLa cells and the use of AICAR in HEK-293 cells promoted phosphorylation of Ser588, but only weak Ser341 and Thr642 phosphorylations and binding to 14-3-3s. Paradoxically however, phenformin activated AMPK without promoting AS160 phosphorylation. The IGF-1-induced phosphorylation of the novel phosphorylated Ser666-Pro site was suppressed by AICAR, and by combined mutation of a TOS (mTOR signalling)-like sequence (FEMDI) and rapamycin. Thus, although AS160 is a common target of insulin, IGF-1, EGF, PMA and AICAR, these stimuli induce distinctive patterns of phosphorylation and 14-3-3 binding, mediated by at least four protein kinases.
Keywords: 14-3-3, Akt/protein kinase B (PKB), Akt substrate of 160 kDa (AS160), GTPase-activating protein (GAP), p90 ribosomal S6 kinase (RSK), serum- and glucocorticoid-induced protein kinase (SGK)
Abbreviations: ACC, acetyl-CoA carboxylase; AGC kinase, protein kinase A/protein kinase G/protein kinase C-family; AICAR, 5-amino-4-imidazolecarboxamide1-β-D-ribofuranoside; AMPK, AMP-activated protein kinase; AS160, Akt substrate of 160 kDa; DSP, dithiobis(succinimidyl propionate); 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; GAP, GTPase-activating protein; GLUT4, glucose transporter 4; GST, glutathione S-transferase; GSV, GLUT4 storage vesicle; HA, haemagglutinin; HEK-293, human embryonic kidney-293; IGF-1, insulin-like growth factor-1; IRAP, insulin-responsive aminopeptidase; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; PAS, phospho-Akt substrate; PDK1, phosphoinositide-dependent kinase 1; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B (also known as Akt); PKC, protein kinase C; PP2A, protein phosphatase 2A; pSer, phosphorylated serine; p70S6K, p70 S6 kinase; PTB, phosphotyrosine binding domain; pThr, phosphorylated threonine; Rheb, Ras enriched in brain; RSK, p90 ribosomal S6 kinase; SGK, serum- and glucocorticoid-induced protein kinase; TORC1, mTOR–raptor (regulatory associated protien of mTOR) complex; TOS motif, mTOR signalling motif; TSC, tuberous sclerosis complex
INTRODUCTION
14-3-3s are C-shaped proteins that dock on to specific phosphorylated sites, forcing changes in the conformation of their targets and/or their interactions with other molecules [1]. Hundreds of proteins that bind to 14-3-3s directly, or as components of multi-subunit complexes, have been purified from mammalian cells using their affinity for 14-3-3s [2–5]. Thus binding of 14-3-3s to phosphoproteins is a common mechanism for linking signalling pathways to the regulation of cellular processes.
A sense of the global regulation of 14-3-3–target interactions can be gleaned by using far Western (14-3-3 overlay) assays to observe how the patterns of phosphorylated 14-3-3-binding proteins change in extracts of cells stimulated in different ways [5,6]. Such experiments show that the phosphorylation of a substantial subset of 14-3-3 targets is promoted by the PI3K (phosphoinositide 3-kinase) pathway, including cardiac PFK-2 (phosphofructokinase 2) [7], TSC2 (tuberous sclerosis complex 2) [8,9], p27Kip1 [10], FOXO3a (forkhead box O3a) [11] and AS160 (Akt substrate of 160 kDa)/TBC1D4 [5,12,13].
AS160/TBC1D4 has been implicated in mediating the translocation of GLUT4 (glucose transporter 4) to the plasma membrane in response to insulin [14], a process whose deregulation occurs early in the pathophysiology of insulin resistance and type 2 diabetes [15]. The emerging role of AS160 in insulin-stimulated GLUT4 trafficking is as follows. (i) In the basal state, most GLUT4 is in intracellular GSVs (GLUT4 storage vesicles). AS160 is a RabGAP (Rab GTPase-activating protein) that promotes hydrolysis of GTP to GDP by Rabs 8A, 10 and 14 on the GSV [16–19]. It is proposed [19–21] that in their inactive GDP-bound state the GSV-bound Rabs inhibit GLUT4 exocytosis. (ii) Upon insulin stimulation, AS160 is phosphorylated [16,22], which leads to its binding to 14-3-3 proteins [13] and the inactivation of the RabGAP activity of AS160 and/or its dissociation from GSVs [17,23]. Thus GSV-associated Rabs are thought to become loaded with GTP and promote events that lead to expression of GLUT4 at the cell surface, thereby mediating the influx of glucose.
The anti-diabetic drug metformin activates the LKB1/AMPK (AMP-activated protein kinase) signalling pathway [24–26], which has an insulin-sensitizing effect on the liver leading to decreased hepatic glucose production [25], and also enhances glucose uptake into tissues, possibly by altering the kinetics of GLUT4 trafficking to and from the plasma membrane. AICAR (5-amino-4-imidazolecarboxamide-1-β-D-ribofuranoside), which also activates AMPK, has been reported to promote phosphorylation of AS160 [27,28], raising the question of whether metformin also regulates AS160. Other PI3K-independent pathways have also been reported to regulate AS160 [28].
Together, these findings suggest that AS160 may be a convergence point for signalling pathways that promote GLUT4 translocation. The tissue distribution of AS160 is wider than GLUT4 however, suggesting that AS160 may also regulate trafficking of vesicles carrying other cargoes. Moreover, although AS160 is subject to multisite phosphorylation [22], most regulatory studies have focussed on Thr642, whose phosphorylation is linked to insulin-mediated 14-3-3 binding [13,17] and GLUT4 trafficking [22], and/or monitored phosphorylation of AS160 using the PAS (phospho-Akt substrate) antibody, which recognizes sequences phosphorylated on a generic Akt/PKB (protein kinase B) phosphomotif [27–30]. In the present study, our initial aim was to define further mechanistic details of 14-3-3 binding to AS160. Our findings led to a wider study in which we have shown that IGF-1 (insulin-like growth factor-1), EGF (epidermal growth factor), PMA and AICAR induce distinct patterns of multisite phosphorylation and 14-3-3 binding of AS160, involving at least four protein kinases.
EXPERIMENTAL
Materials
Synthetic peptides were from Graham Bloomberg (Department of Biochemistry, University of Bristol, Bristol, U.K.), oligonucleotides were from MWG-Biotech, IGF-1 was from Biosource, microcystin-LR was from Linda Lawton (School of Life Sciences, The Robert Gordon University, Aberdeen, Scotland, U.K.), frozen human HeLa cell pellets were from 4c Biotech, Vivaspin concentrators were from Vivascience, tissue culture reagents were from Life Technologies, protease-inhibitor cocktail tablets (catalogue number 1697498) and sequencing-grade trypsin were from Roche Molecular Biochemicals, AICAR was from Toronto Research Chemicals, precast SDS/polyacrylamide gels were from Invitrogen, and DSP [dithiobis(succinimidyl propionate)] was from Perbio. Protein G–Sepharose and other chromatographic matrices were from GE Healthcare. All other chemicals were from BDH Chemicals or Sigma–Aldrich.
Antibodies and protein kinases
Sheep anti-HA (haemagglutinin) was raised against the peptide YPYDVPDYA, and sheep anti-AS160 was raised against KAKIGNKP (near the C-terminus of human AS160; see Supplementary Figure 1 at http://www.BiochemJ.org/bj/407/bj4070231add.htm). Phosphospecific antibodies were raised against the following phosphopeptides: FRSRCSpSVTGVQR [residues 311–324, where pS represents phosphorylated Ser318 (pSer)]; CPRRRHApSAPSHVQ (cysteine+residues 335–347, pSer341); CKAKRSLpTSSLENI [cysteine+residues 562–574, where pT is phosphorylated Thr568 (pThr)]; CKRSLTSpSLENIFS (cysteine+residues 564–576, pSer570); CMRGRLGpSVDSFER (cysteine+residues 582–594, pSer588); CFRRRAHpTFSHPPS (cysteine+residues 636–648, pThr642); CAQGVRpSPLLRQS (cysteine+residues 661–672, pSer666); and GRKRTSpSTCSNES (residues 745–757, pSer751). The pSer318 and pSer751 peptides were conjugated via their N-termini, whereas the others were coupled via the added cysteine residues. Peptides were coupled separately to BSA and keyhole-limpet haemocyanin, mixed and injected into sheep at Diagnostics Scotland. The antibodies were affinity-purified on phosphopeptide–Sepharose columns, and in some cases were also passed through columns coupled to the cognate unphosphorylated peptides and the flow-throughs collected. In each case, the antibodies recognized the synthetic phosphopeptide immunogen, but not the corresponding unphosphorylated peptide in dot blot assays (results not shown). Anti-phosphoERK1/2 (extracellular-signal-regulated kinase; pThr202/pTyr204), anti-pThr308-PKB and anti-PKB/Akt were from Cell Signaling Technology. Western blot analysis used the indicated antibodies at a concentration of 1 μg/ml. Blots in Figure 1, all K19 Western blots and blots of cell lysates were visualized by ECL (enhanced chemiluminescence) reagent, whereas other Western blots used secondary antibodies that were detected using the Odyssey Infrared Imaging System (LI-COR). 14-3-3 overlays [using DIG (digoxigenin)-labelled 14-3-3 in place of a primary antibody) used ECL detection [5].
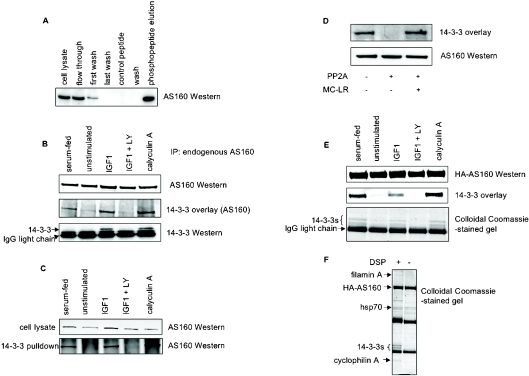
Figure 1. Phosphorylation-dependent binding of AS160 to 14-3-3 proteins in response to IGF-1.
(A) Isolation of AS160 by 14-3-3-affinity chromatography of HeLa cell extracts. Crude extract, flow through, first wash fractions (40 μg) and 5 μl of the concentrated last wash, control peptide and final wash fractions (<1 μg protein) from the 14-3-3 column were loaded on to a 4–12% Bis/Tris SDS gel, alongside 2 μg of the proteins that were eluted with the 14-3-3-binding phosphopeptide, ARAApSAPA (phosphopeptide elution). The gel was transferred on to nitrocellulose, and anti-AS160 Western blot analysis was performed. Protein stains and 14-3-3 overlay signals for this column were similar to those reported in [5]. (B) Untransfected HEK-293 cells that had been serum-fed, unstimulated, IGF-1-stimulated, IGF-1-stimulated in the presence of LY294002 (LY) and treated with calyculin A were lysed in the presence of DSP cross-linker. Endogenous AS160 was immunoprecipitated from lysates (6 mg), denatured and run on a 4–12% Bis/Tris SDS gel, transferred on to nitrocellulose, and the 14-3-3-binding ability of AS160 was tested by 14-3-3 overlay (middle panel). Cellular 14-3-3 proteins that co-precipitated with endogenous AS160 were identified by Western blot analysis with pan-14-3-3 antibodies (K-19 from Santa Cruz Biotechnology; bottom panel). (C) 14-3-3–Sepharose (20 μl, prepared as described in [5]) was incubated with HEK-293 cell lysate (6 mg) from cells that were serum-fed, unstimulated (serum-starved), IGF-1-stimulated, IGF-1-stimulated in the presence of LY294002 (LY) and treated with calyculin A. The 14-3-3–Sepharose was washed three times in a high-salt buffer [25 mM Tris/HCl (pH 7.5 at 4 °C), 25 mM NaF and 500 mM NaCl], followed by two washes in a low-salt buffer [25 mM Tris/HCl (pH 7.5 at 4 °C), 25 mM NaF and 150 mM NaCl]. AS160 bound to the 14-3-3–Sepharose was identified by Western blot analysis. (D) Endogenous AS160 was immunoprecipitated from 4 mg of HEK-293 cell extract, and incubated for 30 min at 30 °C in the presence of 50 m-units/ml PP2A, with or without the inhibitor microcystin-LR (MC-LR; 5 mM), as indicated. Proteins were separated by SDS/PAGE, transferred on to nitrocellulose and the AS160 was probed for binding to 14-3-3s in the overlay assay. (E) HA-tagged AS160A (see the Experimental section) was immunoprecipitated from lysates (3 mg) of transfected HEK-293 cells that had been serum-fed, unstimulated, IGF-1-stimulated, IGF-1-stimulated in the presence of LY294002 (LY) and calyculin A treated. The immunoprecipitated protein was analysed by the 14-3-3 overlay. In addition, HA–AS160 was immunoprecipitated from cell lysates (1 mg), denatured in LDS (lithium dodecyl sulfate) sample buffer (Invitrogen) and run on a 4–12% Bis/Tris SDS gel, which was stained with Colloidal Coomassie Blue (bottom panel). (F) HA–AS160A was immunoprecipitated from cell lysates (1 mg) prepared with and without DSP cross-linker, as indicated. The immunoprecipitated protein was run on a 4–12% Bis/Tris SDS gel, which was stained with Colloidal Coomassie Blue, and the proteins indicated were identified by tandem MS analysis of tryptic digests. (See the text and Supplementary Table 1 at http://www.BiochemJ.org/bj/407/bj4070231padd.htm for details.) Hsp70, heat-shock protein 70.
Purified recombinant protein kinases, generated in the DSTT (Division of Signal Tranduction Therapy, University of Dundee, Dundee, Scotland, U.K.), were His–PKBα-S473D (p90 ribosomal S6 kinase 1) (residues 118–480 of human protein) expressed in insect cells and activated by PDK1 (phosphoinositide-dependent kinase 1); His–RSK1 (p90 ribosomal S6 kinase 1) (residues 1–735 of rat protein) expressed in insect cells and activated by p42MAPK (mitogen-activated protein kinase) and PDK1; SGK1 (serum- and glucocorticoid-induced protein kinase 1):-S422D (residues 60–431 of human protein) expressed in insect cells and activated by PDK1; and bacterially expressed GST–AMPK-T172D (residues 3–308 of rat protein; where GST is glutathione S-transferase). AMPK was purified from rat liver by Kevin Green (Division of Molecular Physiology, College of Life Sciences, University of Dundee, Dundee, Scotland, U.K.). Unless indicated, the protein kinases were used at 1 unit/ml, where 1 unit is the nmol of phosphate incorporated per min at 30 °C into the substrate peptides Crosstide (GRPRTSSFAEG) for PKB and SGK1, Long-S6 (KEAKEKRQEQIAKRRRLSSLRASTSKSGGSQK) for RSK1 and AMARA peptide (AMARAASAAALARRR) for AMPK.
Molecular biology: splice variants of AS160
Two forms of AS160 appear to be products of one locus (GenBank® accession number NM_014832; gene id: ENSG00000136111). One form (termed AS160B) is identical with Z25171 from a skeletal muscle cDNA library. A shorter form (AS160A), more commonly represented in the sequence databases, is identical with AS160B except for the deletion of residues 679–733 encoded by exon 11. Constructs to generate HA–AS160A (pCMV5.HA AS160A) and HA–AS160B (pCMV5.HA AS160B) were generated by RT (reverse transcriptase)-PCR amplification from RNA extracted from HeLa cells and human skeletal muscle respectively. Bacterial expression plasmids for GST–AS160 fusions (pGEX6P AS160A and pGEX6P AS160B) were generated by standard procedures. Site-directed mutagenesis used the QuikChange® protocol (Stratagene) and KOD HotStart DNA polymerase (Novagene). Sequences of DNA constructs were checked (www.dnaseq.co.uk).
Results are shown for the HA–AS160A and GST–AS160A forms, although experiments in Figure 7 were repeated with AS160B and gave similar results (results not shown). Features of AS160 and the related TBC1D1 are shown in Supplementary Figure 1 (see http://www.BiochemJ.org/bj/407/bj4070231add.htm). Unless stated, residue numbers given correspond to AS160B.
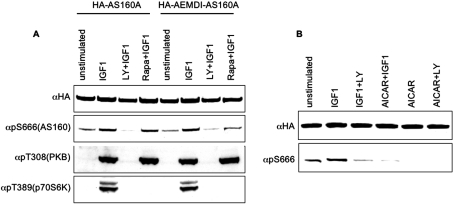
Figure 7. Effect of the FEMDI to AEMDI mutation, rapamycin and AICAR on phosphorylation of Ser666.
(A) HEK-293 cells transfected with HA–AS160A and HA–AEMDI-AS160A were stimulated with IGF-1, in the presence or absence of LY294002 (LY) and rapamycin (Rapa), as indicated. Crude lysates (60 μg) were probed with antibodies against pThr308-PKB and pThr389-p70S6K. HA-tagged proteins were immunoprecipitated, run on SDS/PAGE, transferred on to nitrocellulose and probed with the antibodies indicated. (B) HEK-293 cells transfected with HA–AS160A were stimulated with IGF-1, in the presence and absence of AICAR, as indicated. HA-tagged proteins were immunoprecipitated, run on SDS/PAGE, transferred on to nitrocellulose and probed with the antibodies indicated.
Cell stimulations, cross-linking, lysis and immunoprecipitations
Human HEK (human embryonic kidney)-293 cells cultured on 10-cm-diameter dishes in medium containing 10% (v/v) serum were used untransfected, or 16–30 h after transfection with the indicated plasmids. Cells were serum-starved for a further 4–12 h (unstimulated), then stimulated for 20 min with 50 ng/ml IGF-1. Where indicated, calyculin A was used at 100 nM for approx. 3 min, AICAR at 2 mM for 60 min, phenformin at 2 mM for 60 min, EGF at 50 ng/ml for 15 min and PMA at 100 ng/ml for 30 min. Where indicated, cells were incubated with LY294002 (100 μM for 1 h), wortmannin (100 nM for 1 h), U0126 (10 μM for 1 h), rapamycin (50 nM for 30 min), Go6983 (1 μM for 30 min) and BI-D1870 (10 μM for 30 min) prior to stimulation with IGF-1 and other stimuli. After stimulation, cells were lysed in 0.2 ml of ice-cold lysis buffer comprising 25 mM Tris/HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM benzamidine, 0.2 mM PMSF, 0.1% (v/v) 2-mercaptoethanol, 1 μM MC-LR, 0.27 M sucrose and one mini Complete™ proteinase inhibitor cocktail tablet (Roche) per 10 ml of lysis buffer [5]. For experiments using a chemical cross-linker, cells were rinsed with ice-cold PBS, lysed in 0.3 ml of lysis buffer containing DSP (2.5 mg/ml from 250 mg/ml in DMSO) for 30 min on ice, and unreacted cross-linker was quenched with 75 μl of 1 M Tris/HCl (pH 7.4) with a further 30 min incubation [31].
For immunoprecipitations with anti-AS160 and anti-HA, 4 μg of antibody/mg of lysate was mixed at 4 °C for 1 h, then Protein G–Sepharose (30 μl of a 50% suspension in lysis buffer) was added and mixed for a further 1 h. The suspension was centrifuged at 12000 g for 1 min between washes.
The human HeLa cell lines stably transfected with wild-type LKB1 and kinase-dead LKB1 were those derived by Sapkota et al. [32].
RESULTS
IGF-1 promotes the phosphorylation and binding of AS160 to 14-3-3 proteins
Previously, AS160 was isolated from HeLa cell extracts by 14-3-3-affinity chromatography [5]. Western blot analysis confirmed that AS160 bound to 14-3-3–Sepharose and was specifically eluted by a canonical 14-3-3-binding phosphopeptide, ARAApSAPA, but not a control peptide (Figure 1A).
When endogenous AS160 was immunoprecipitated from extracts of HEK-293 cells that had been stimulated with serum, IGF-1 or the protein phosphatase inhibitor calyculin A, the isolated AS160 bound directly to 14-3-3 proteins in an overlay assay (Figure 1B, middle panel). There was only a trace 14-3-3-binding signal for AS160 from unstimulated cells, or cells stimulated with IGF-1 in the presence of the PI3K inhibitor LY294002 (Figure 1B, middle panel). Similarly, AS160 bound to 14-3-3–Sepharose in extracts of cells that had been stimulated with serum, IGF-1 and calyculin A (Figure 1C, lower panel). The IGF-1-induced 14-3-3-binding signal was unaffected by the mTOR (mammalian target of rapamycin) inhibitor rapamycin or U0126, which inhibits the activation of the classical MAPK pathway (results not shown).
In contrast with the 14-3-3 overlays, only small sub-stoichiometric amounts of 14-3-3 proteins were detected in anti-AS160 immunoprecipitates from IGF-1-stimulated cells (results not shown). However, 14-3-3 proteins were co-immunoprecipitated with AS160 from IGF-1- or calyculin A-stimulated cells when protein–protein interactions were stabilized by DSP, which reversibly links primary amines (Figure 1B, bottom panel). The co-precipitation of 14-3-3 proteins with AS160 was specifically induced by IGF-1 and calyculin A, and blocked by LY294002 (Figure 1B, bottom panel).
The ability of AS160 from IGF-1-induced cells to bind to 14-3-3 proteins was abolished by dephosphorylation of the AS160 with PP2A (protein phosphatase 2A) in vitro, and this was prevented by the PP2A inhibitor microcystin-LR (Figure 1D).
IGF-1 also induced the binding of 14-3-3 proteins to HA–AS160 in transfected cells, as determined by binding of the extracted HA–AS160 to 14-3-3 proteins in the overlay assay (Figure 1E, middle panel), and co-precipitation of 14-3-3 proteins (Figures 1E, bottom panel, and 1F). MS analysis of tryptic digests showed that at least six of the seven human 14-3-3 isoforms were bound to the HA–AS160 extracted from IGF-1-stimulated cells in the presence of DSP (see Supplementary Table 1 at http://www.BiochemJ.org/bj/407/bj4070231add.htm). The upper Coomassie-Blue-stained band (Figure 1F) contained peptides that are unique to the 14-3-3ϵ. The lower band (Figure 1F) contained peptides specific to 14-3-3θ, γ, α/β, η and δ/ζ. No peptides specifying 14-3-3σ were identified.
Identification of phosphorylated sites on AS160, including known sites and the novel pSer666-Pro site
Digests of endogenous AS160 were analysed by liquid chromatography MS [33] to identify IGF-1-induced phosphorylations. Initially, precursor ion scans were run to detect phosphopeptides (Figure 2, and results not shown). An MRM (multiple reaction monitoring) analysis was then performed to look for the known phosphorylated sites on AS160 (see Supplementary Table 2 at http://www.BiochemJ.org/bj/407/bj4070231add.htm) [22]. In addition, pSer318 and pThr642 (also identified in [22]) were detected in endogenous AS160 using phosphospecific antibodies (see below), and by mass spectral analysis of recombinant AS160 isolated from human cells (results not shown).
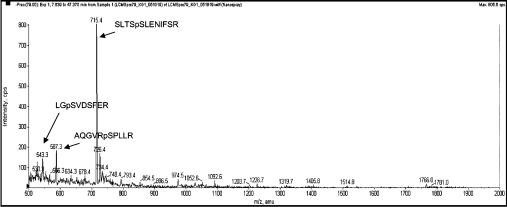
Figure 2. Precursor ion scan for the tryptic digest of endogenous AS160 from calyculin A-treated cells.
Using liquid chromatography MS on an Applied Biosystems 4000 Q-TRAP, a precursor ion scan identifies those parent masses that yield daughter ions with an m/z of 79 kDa (PO3−), which is only the phosphopeptides indicated. The phosphorylated peptides were analysed further to identify the phosphorylated residues (see Supplementary Table 2 at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
In addition to known sites, a new phosphopeptide from endogenous AS160 was identified by tandem MS as AQGVRpSPLLR (Figure 2). The pSer666 within this phosphopeptide lies within a pSer-Pro motif that is not a consensus site for PKB. The MS data indicated a marked increase in phosphorylation at Ser666 in response to IGF-1, and the increased phosphorylation was blocked by LY294002 (see Supplementary Table 3 at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
Mutation analysis indicates that 14-3-3 binds to pThr642 and pSer341
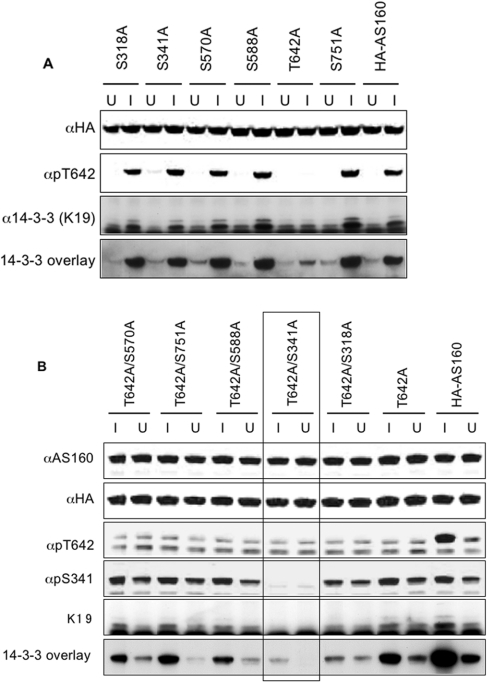
HA–AS160 with each of the seven phosphorylation sites individually mutated from serine/threonine to alanine was tested for 14-3-3 binding using overlay assays and by co-precipitation of 14-3-3 proteins with HA–AS160 from cell extracts. Only the Thr642Ala mutation had a marked effect in preventing the IGF-1-induced binding of 14-3-3 proteins to HA–AS160 (Figure 3A).
Figure 3. Identification of 14-3-3-binding sites on AS160.
(A) HEK-293 cells growing in the presence of serum were transfected with vectors to express wild-type HA–AS160A and mutants with the single serine/threonine to alanine mutations indicated. The cells were unstimulated (U) or stimulated with IGF-1 (I), and HA-tagged proteins were immunoprecipitated from 500 μg of extract, and subjected to anti-HA, anti-pThr642-AS160 and anti-14-3-3 (K19) Western blot analysis, and 14-3-3 overlays. (B) HEK-293 cells transfected with wild-type HA–AS160A, Thr642Ala-AS160A and double mutants were treated with IGF-1 (I) or unstimulated (U). The HA-tagged proteins were immunoprecipitated from 500 μg of cell extract, one-fifth of each immunoprecipitate was loaded on to each lane for immunoblotting, and blots were subject to anti-AS160, anti-HA, anti-pThr642-AS160, anti-pSer341 and anti-14-3-3 (K19) Western blot analysis, and 14-3-3 overlays. The specificity of the phosphospecific antibodies is demonstrated in Supplementary Figure 2(A) (at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
The 14-3-3 binding of Thr642Ala–AS160 was not completely abolished however (Figure 3A), indicating the existence of a second lower-affinity 14-3-3-binding site. Therefore double mutants of Thr642Ala combined with serine to alanine mutations of each of the other phosphorylated sites were generated. Of the double mutants, the combination of Thr642Ala and Ser341Ala completely prevented the IGF-1-induced binding of 14-3-3 proteins to AS160 (boxed lanes in Figure 3B). Consistent with phosphorylation of Ser341 being responsible for the basal binding of 14-3-3 proteins to AS160, the Ser341Ala mutation abolished the binding of 14-3-3 proteins to AS160 in unstimulated cells (Figure 3B, and results not shown).
In combination with Thr642Ala, the mutation of Ser318 to an alanine residue also decreased the binding of 14-3-3 proteins to AS160 (Figure 3B), consistent with the Ser318Ala mutation causing a decrease in the phosphorylation of Ser341 (Figure 3B and Supplementary Figure 2A at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
Generation of phosphospecific AS160 antibodies to monitor the phosphorylation of seven sites on AS160
To examine the cellular regulation of these phosphorylations further, we raised phosphospecific antibodies against each of the seven phosphorylated residues on AS160 (pSer318-AS160, pSer341-AS160, pSer570-AS160, pSer588-AS160, pThr642-AS160, pSer666-AS160 and pSer751-AS160; see Supplementary Figure 2A at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
The recognition of AS160 by anti-PAS was largely abolished by the Thr642Ala mutation, indicating that pThr642 is the prime anti-PAS recognition site on AS160 from cells stimulated with serum, IGF-1 and insulin (see Supplementary Figure 2B at http://www.BiochemJ.org/bj/407/bj4070231add.htm and results not shown).
Phosphorylation of AS160 in response to IGF-1 and AICAR, but not phenformin, in HEK-293 cells
The phosphospecific antibody data for endogenous AS160, and HA–AS160 from transfected cells, showed that the phosphorylation of each of the seven sites was enhanced when cells were stimulated with serum and IGF-1. These phosphorylations were prevented by LY294002 (Figure 4A, and results not shown), except for pSer588 whose phosphorylation was partly inhibited by LY294002 (Figure 4A, and results not shown). There was a high basal phosphorylation of Ser341 and Ser666 in unstimulated cells, which was also abolished by LY294002 (Figure 4A).
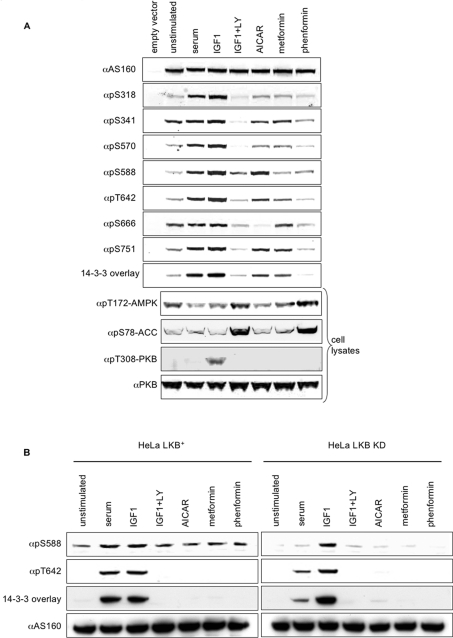
Figure 4. Phosphorylation of seven sites on AS160 in response to different stimuli in HEK-293 cells, and the effect of LKB1 on the phosphorylation of Ser588.
(A) HEK-293 cells transfected with HA–AS160A were stimulated as indicated. Crude lysates (60 μg per lane) were probed for binding to the antibodies as indicated along the left-hand side of the upper panels. The cells transfected with empty vector were serum-starved before harvesting. In the lower four panels, extracts (40 μg) of HEK-293 cells that had been stimulated as indicated were analysed by Western blotting with antibodies to monitor the phosphorylation status of the activation loop on AMPK, the AMPK site on ACC (pSer79 in the rat protein, pSer80 in the human), and the activating (pThr308) phosphorylation site on PKB. (B) Endogenous AS160 was immunoprecipitated from HeLa cells stably transfected with either LKB1 or kinase-dead (KD) LKB1 [32], and Western blot analysis and 14-3-3 overlays were performed, as indicated. LY, LY294002.
The AMPK activator AICAR promoted phosphorylation of Ser588 of AS160 (Figure 4A), with much weaker phosphorylations of Ser341, Thr642 and Ser751 (Figure 4A and see below). Consistent with the much lesser phosphorylation of Ser341 and Thr642 compared with that seen with IGF-1, AICAR promoted only weak binding of 14-3-3 proteins to AS160 that was more easily detected in the overlay assay (Figure 4A) than by co-immunoprecipitation of AS160 with 14-3-3 proteins from cell extracts (results not shown).
Phenformin promoted a much stronger phosphorylation of Thr172 in the activation loop of AMPK, and of the AMPK target ACC (acetyl-CoA carboxylase), compared with AICAR and metformin (Figure 4A). Despite its strong activation of AMPK however, phenformin did not stimulate phosphorylation of AS160 (Figure 4A). Metformin promoted only weak phosphorylation of several sites on AS160 (Figure 4A), and, in particular, metformin did not stimulate phosphorylation of Ser588 to the same extent as AICAR (Figure 4A). We also noticed that phosphorylation of both AMPK and ACC was strongly promoted by LY294002 in HEK-293 cells (Figure 4A), but not in other cells tested (results not shown). Neither phenformin nor metformin inhibited any of the IGF-1- and AICAR-stimulated phosphorylations of AS160 in HEK-293 cells (results not shown).
In view of the paradoxical effects of AICAR and phenformin, we examined whether LKB1, an upstream activator of AMPK had any role in regulating phosphorylation of AS160. LKB1 is absent from HeLa cells [32] and AICAR did not promote phosphorylation of AS160 in these cells (results not shown). In HeLa cells that were stably transfected with wild-type LKB1, the presence of the active LKB1 was itself sufficient to induce phosphorylation of Ser588 of endogenous AS160, even in the absence of any AMPK activators (Figure 4B, left-hand panels). Kinase-dead LKB1 did not induce phosphorylation of AS160 at Ser588 (Figure 4B, right-hand panels).
In vitro phosphorylation of GST–AS160 by PKB, AMPK, SGK1 and RSK1: identification of an eighth phosphorylation site at Thr568
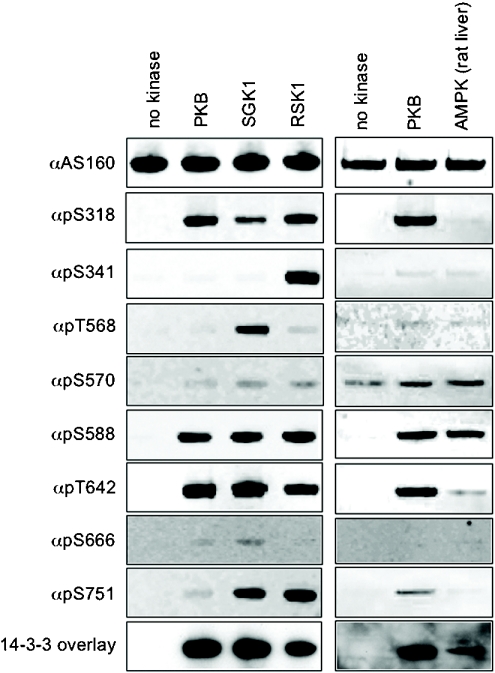
Consistent with its proposed role in phosphorylation of AS160 [14], PKB preferentially phosphorylated Ser318, Ser588 and Thr642, and also catalysed trace phosphorylation of Ser570 and Ser751 in vitro (Figure 5). In contrast, AMPK preferentially phosphorylated Ser588 of AS160, phosphorylated Ser570 to a similar degree, as did PKB, and phosphorylated Thr642 to a lesser extent (Figure 5, right-hand panels). Compared with PKB, other AGC (protein kinase A/protein kinase G/protein kinase C-family) kinases displayed distinct signatures of AS160 phosphorylation in vitro (Figure 5, left-hand side and results not shown). In common with PKB, SGK1 and RSK1 phosphorylated Ser318, Ser588 and Thr642 (Figure 5, left-hand panels). In addition, RSK1 also phosphorylated Ser341 and Ser751; and SGK phosphorylated Ser751. MS analysis of tryptic digests of in vitro-phosphorylated AS160 also revealed a new phosphorylated site at Thr568, within the sequence KAKRSLpT568SSLENI, two residues N-terminal to the pSer570 site (results not shown). A phosphospecific pThr568-AS160 antibody was raised and tested for phospho- and sequence specificity (see Supplementary Figure 2C at http://www.BiochemJ.org/bj/407/bj4070231add.htm), and using this antibody it was found that SGK1 was the best Thr568 kinase of those tested, although this site was also phosphorylated by RSK1 (Figure 5, left-hand panels). These kinases displayed no or barely detectable phosphorylation of Ser666 (Figure 5).
Figure 5. In vitro phosphorylation of AS160 by AMPK and the AGC kinases, PKB, SGK1 and RSK1.
In the right-hand panels, GST–AS160A was expressed and purified from Escherichia coli, and phosphorylated in vitro with PKB (at 1 unit/ml) and AMPK purified from rat liver (10 units/ml). The phosphorylated proteins (300 ng per lane) were analysed by Western blotting using phosphospecific AS160 antibodies (see Supplementary Figures 3A and 3C at http://www.BiochemJ.org/bj/407/bj4070231add.htm) and 14-3-3 overlay, as indicated. Similarly, in the left-hand panels, GST–AS160A was phosphorylated in vitro by PKB, SGK1 and RSK1 (at 1 unit/ml).
Phosphorylation and 14-3-3 binding of AS160 in response to EGF and PMA in HEK-293 cells: a role for RSK in mediating the response to PMA
We next assessed whether Thr568 of AS160 is phosphorylated in cells, and whether RSK is a physiological AS160 kinase. Alongside IGF-1, AICAR and phenformin, cells were treated with EGF and the phorbol ester PMA, which stimulate RSK isoforms via the MAPK family members ERK1 and ERK2 ([34] and references within) (Figure 6 and see Supplementary Figure 3 at http://www.BiochemJ.org/bj/407/bj4070231add.htm).
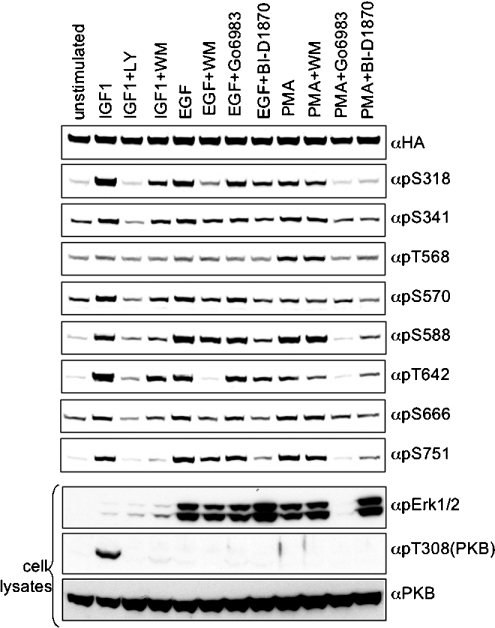
Figure 6. Phosphorylation of all eight sites on AS160 in response to IGF-1, EGF and PMA in HEK-293 cells, and effects of protein kinase inhibitors.
HEK-293 cells transfected with HA–AS160A were stimulated after pre-exposure to inhibitors, as indicated, and HA–AS160A was analysed by Western blotting. In addition, cell lysates (60 μg per lane) were analysed by Western blotting to detect phosphorylation of ERK1/2, pThr308 of PKB and total PKB. LY, LY294002; WM, wortmannin.
HA–AS160 and 14-3-3 proteins were co-immunoprecipitated, and AS160 was phosphorylated on several sites and could bind to 14-3-3 proteins in overlay assays, when extracted from cells stimulated with serum, IGF-1, EGF and PMA (Figure 6 and see Supplementary Figure 3 at http://www.BiochemJ.org/bj/407/bj4070231add.htm). Of these stimuli, PMA gave the weakest 14-3-3-binding signals for AS160, consistent with its lesser stimulation of Thr642 phosphorylation (Figure 6 and Supplementary Figure 3). In contrast, PMA induced the strongest phosphorylation of Thr568, showing that this site was phosphorylated in cells (Figure 6) as well as in vitro (Figure 5).
When PMA-induced and PKC (protein kinase C)-mediated phosphorylation of ERK1 and ERK2 was blocked by Go6983 (Figure 6) [35], this inhibitor largely abolished phosphorylation of AS160 in response to PMA (Figure 6). In addition, the RSK inhibitor BI-D1870 [34] markedly suppressed the responses to PMA, except the weak Thr642 phosphorylation (Figure 6). Go6983 and BI-D1870 also inhibited PMA-induced binding of AS160 to 14-3-3 proteins (results not shown). Wortmannin had no effect on PMA-induced phosphorylation of AS160 (Figure 6).
In contrast, wortmannin inhibited EGF-induced phosphorylation of Ser318 and Thr642, with little or no effect on the other EGF-regulated sites (Figure 6), although the RSK inhibitor BI-D1870 inhibited all of the EGF-stimulated phosphorylations to different extents (Figure 6). In contrast with PMA, Go6983 inhibited phosphorylation of neither ERK1/2 nor AS160 in response to EGF.
As described previously, the IGF-1-stimulated phosphorylations were blocked by LY294002, and largely inhibited by wortmannin (Figure 6).
IGF-1-induced phosphorylation of Ser666 is partially inhibited by combined mutation of a TOS (mTOR signalling)-like motif in AS160 and rapamycin, and is blocked by AICAR
The IGF-1-induced phosphorylation of the novel pSer666-Pro site was not affected by SB203580, a specific inhibitor of SAPK (stress-activated protein kinase) 2a/p38, or U0126, an inhibitor of ERK1/2 activation (results not shown).
mTOR mediates the insulin- and IGF-1-induced phosphorylation of pSer/Thr-Pro motifs on 4E-BP1 (eukaruotic initiation factor 4E-binding protein 1)/PHAS (pT37PGGTLFSTpT46P) [36] and p70S6K (p70 S6 kinase; PVDS371P) [37]. TOS motifs on these proteins act as docking sites for the TORC1 [mTOR–raptor (regulatory associated protein of mTOR) complex], and are required for the phosphorylation of the Ser/Thr-Pro sites [38]. Intriguingly, the AS160 protein contains a sequence, FEMDI, identical with the TOS motif on 4E-BP1. Consistent with a functional role for this motif, when the FEMDI motif was mutated to AEMDI the IGF-1-stimulated phosphorylation of Ser666 was more inhibited by rapamycin, compared with the wild-type HA–AS160 (Figure 7A, last lane). However, neither rapamycin alone nor mutation of the FEMDI motif alone had any obvious effect on Ser666 phosphorylation in IGF-1-stimulated cells (Figure 7A). As expected, the IGF-1-induced phosphorylation of a TOS-dependent site on p70S6K was prevented by rapamycin alone (Figure 7A, bottom panel).
Among other effectors tested, the AMPK activator AICAR abolished both basal and IGF-1-stimulated phosphorylation of Ser666 of AS160 (Figure 7B).
DISCUSSION
In the present study we identified a specific and reversible interaction between phosphorylated AS160 and 14-3-3 proteins in cells stimulated with IGF-1. The affinity of the phosphorylated AS160–14-3-3 interaction is sufficient for the phosphorylated AS160 to bind rapidly to the 14-3-3 proteins inside the cell. However, it is poised at a point where the phosphorylated AS160 and 14-3-3 proteins dissociate during the washing procedures of immunoprecipitation, unless stabilized by cross-linking. Such reversibility makes physiological sense for a regulatory interaction that must respond dynamically to changes in the stimulation and metabolic status of the cell. Cellular AS160 binds to at least six of the seven human isoforms of 14-3-3 proteins (Figure 1F and Supplementary Table 1 at http://www.BiochemJ.org/bj/407/bj4070231add.htm). 14-3-3σ was absent from the AS160 immunoprecipitates, and was not detected in cell extracts using anti-14-3-3σ antibodies (results not shown). The 14-3-3 proteins that interact with AS160 may simply reflect the complement of isoforms in this particular cell type, although isoform selectivity cannot be ruled out.
In the present study we identified eight phosphorylated residues in AS160, of which six have been reported previously [22], whereas the pSer666-Pro and pThr568 sites are novel. However, studies of the responsiveness of AS160 to extracellular stimuli generally use the anti-PAS antibody that primarily detects pThr642 ([27,30]; Supplementary Figure 2B at http://www.BiochemJ.org/bj/407/bj4070231add.htm). Our wider analysis shows how distinct patterns of multisite phosphorylation of AS160 are induced by incoming signals from the PI3K pathway, the PKC/ERK/RSK pathway, LKB1/AMPK and other signalling pathways. By comparing the phosphorylation status of the eight sites in response to extracellular stimuli, the effects of protein kinase inhibitors, and the in vitro phosphorylation specificity of protein kinases, we can start to assign candidate AS160 kinases to specific sites. For example, our results are consistent with the possibility that PKB phosphorylates Thr642 in response to IGF-1, but the IGF-1-activated Ser341 kinase has yet to be identified (Figures 4A, 5 and 6). In contrast, RSK is a good candidate for phosphorylating Ser341 in response to PMA (Figures 5 and 6). Further gaps need to be filled in with respect to which protein kinases phosphorylate which sites on AS160 in response to which stimuli. Once that task is advanced further, we suggest that the phosphospecific antibody signatures for AS160 will provide valuable information of the responsiveness of different signalling pathways and protein kinases to cellular stimuli and drugs.
Subtle interdependencies were observed among the phosphorylations. For example, a Ser318Ala mutation decreased phosphorylation of Ser341 (Figure 3B and Supplementary Figure 2A at http://www.BiochemJ.org/bj/407/bj4070231add.htm). In the unstimulated state Ser341 is phosphorylated. Although this looks like a perfect mode I 14-3-3-binding motif (http://scansite.mit.edu/motifscan_seq.phtml), Ser341 phosphorylation mediates only a low-affinity basal interaction with 14-3-3 proteins, which is markedly enhanced when Thr642 is phosphorylated in response to insulin or IGF-1. Consistent with an essential role for 14-3-3 proteins in GLUT4 translocation [13], Thr642 is critical for mediating the translocation of GLUT4 and glucose uptake in response to insulin [22]. We suggest that a single 14-3-3 dimer might bind pSer341 and pThr642. Intriguingly, these sites flank the second PTB (phosphotyrosine-binding domain) on AS160 (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/407/bj4070231add.htm), which was reported to bind to IRAP (insulin-responsive aminopeptidase; [23]) thereby docking AS160 onto the GSVs [17,23]. Thus 14-3-3 binding might disrupt the interaction between IRAP and the second PTB domain of AS160, thereby releasing AS160 from the GSVs, enabling the Rabs to be loaded with GTP to promote translocation of the GSVs towards the cell surface. However, we have thus far failed to observe the IRAP–AS160 interaction either in vitro or in cell extracts (results not shown).
The potential for AMPK to mediate the stimulatory effects of the nucleoside AICAR, as well as exercise, on GLUT4 translocation and glucose uptake into skeletal muscle and adipocytes has been studied [27,29,30] using the anti-PAS antibody which primarily detects pThr642 (see Supplementary Figure 2B at http://www.BiochemJ.org/bj/407/bj4070231add.htm). However, we find that Ser588 is most strongly phosphorylated by AMPK, whereas Thr642 is a secondary site of phosphorylation by this kinase in vitro (Figure 5) and in cell responses to AICAR (Figure 4A). Intriguingly, Sano et al. [22] pinpointed the double Ser588Ala/Thr642Ala-AS160 mutant as having a greater inhibitory effect on insulin-stimulated GLUT4 translocation than the single Thr642Ala-AS160 mutant, suggesting that Ser588 and Thr642 can both influence GLUT4 translocation.
The high pSer588/pThr642 phosphorylation ratio induced by AICAR (Figure 4A), mirrors the specificity of AMPK towards AS160 in vitro (Figure 6), consistent with AMPK mediating the AICAR-induced phosphorylations. AICAR in cells is converted by adenosine kinase into the monophosphorylated nucleotide ZMP, which mimics the effects of AMP on the AMPK system [39]. However, the AICAR-induced phosphorylation of AS160 occurred with barely detectable AMPK phosphorylation and activation (Figure 4A, and results not shown). This lack of effect of AICAR on total AMPK in HEK-293 cells is consistent with previous findings [40]. AMPK is an α/β/γ heterotrimer and, theoretically, twelve forms of AMPK could exist comprising combinations of the two α-, two β- and three γ-subunit isoforms. AICAR activates AMPK α2 complexes in mouse muscle [41], whereas AMPK α2 and γ3 subunits are required for AICAR-induced AS160 phosphorylation detected by the PAS antibody [30]. The α2 subunit of the AMPK, which is a less major form than α1 in HEK-293 cells (D. G. Hardie and S. Hawley, unpublished work), may therefore be the AS160 kinase. If so, one would have to explain how both metformin and phenformin promoted a marked phosphorylation of the AMPK and its downstream target ACC, and yet have little effect on phosphorylation of AS160, in contrast with the effects of AICAR. Metformin and phenformin inhibit Complex I of the mitochondrial respiratory chain and are thought to activate AMPK by increasing the intracellular AMP/ATP ratio [39]. We considered the possibility that AMPK phosphorylates Ser588 of AS160, but that, being metabolic poisons, metformin and phenformin cause another effect that inhibits AS160 phosphorylation. However, neither metformin nor phenformin blocked the IGF-1- or AICAR-induced phosphorylations of AS160. Another possibility is that phenformin activates a phosphatase that can obscure phenformin-induced phosphorylation, but is insufficient to impact on IGF-1- and AICAR-induced phosphorylations. We emphasize that the paradoxical effects of AICAR and phenformin do not seem to be unique to HEK-293 cells. These were also seen for the AS160 from rat L6 myotubes (results not shown), which are commonly used for studying the regulation of glucose trafficking by insulin. However, the majority of extractable AS160 in L6 cells was a truncated form lacking the regulatory phosphorylation sites (results not shown).
LKB1 is the upstream kinase that mediates the activation of AMPK by AICAR and metformin/phenformin [39]. LKB1 also activates a number of AMPK-related protein kinases [39]. We found that Ser588 of AS160 was phosphorylated in HeLa cells containing active LKB1, but not in cells carrying the kinase-dead LKB1 (Figure 4B). Overall, these findings are consistent with AS160 being phosphorylated by AMPK or another LKB1-dependent enzyme. Our findings agree with the lack of effect of metformin on basal or insulin-induced AS160 phosphorylation measured with the PAS antibody [42]. To our knowledge, no other researchers have reported effects of metformin/phenformin on AS160. Perhaps others have made the same findings with AICAR and metformin/phenformin as we report in the present study and have been similarly puzzled by these paradoxical data.
The identity of the kinase(s) that phosphorylates Ser666 is unknown. The IGF-1-induced phosphorylation of Ser666 on AS160 was not blocked by rapamycin, unlike the IGF-1-dependent phosphorylation of pSer-Pro sites on eEF2K (elongation factor 2 kinase), p70S6K and 4E-BP1 ([43]; Figure 7A). Moreover, phosphorylation of Ser666 was not inhibited by mutation of a putative TOS motif (FEMDI), which mediates mTOR/TORC1-dependent phosphorylation of downsteam targets. We cannot rule out a role for mTOR at this stage though, because the combination of mutation of the TOS-like motif and rapamycin together suppressed the IGF-1-induced phosphorylation of Ser666 of AS160. The FEMDI sequence is conserved in AS160 proteins from all other mammalian species currently cited in sequence databases. Moreover, the IGF-1-induced Ser666 phosphorylation was inhibited by AICAR. Activation of mTOR in the TORC1 complex by insulin and IGF-1 initially involves PKB, which inhibits the RhebGAP (where Rheb is Ras enriched in brain) activity of the TSC1/TSC2 (TSC heterodimer), and thereby increases the RhebGTP/RhebGDP ratio [44]. AMPK inhibits insulin-induced mTOR/TORC1 signalling by mechanisms that include phosphorylation and activation of TSC2 [44–47]. Therefore the finding that AICAR blocks IGF-1-induced phosphorylation of Ser666 of AS160 is consistent with a role for the mTOR family.
Recently, a mutation in the TBC1D1 gene, encoding a protein related to AS160, was linked with severe obesity in females [48]. TBC1D1 was isolated as a 14-3-3-binding protein [3]. Thr596 in TBC1D1, analogous to Thr642 in AS160, is phosphorylated in response to insulin in adipocytes [49] and some other AS160 phosphorylation sites are conserved in TBC1D1 (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/407/bj4070231add.htm), and we found that 14-3-3 proteins bind to TBC1D1 in response to IGF-1 and insulin (S. Chen and C. MacKintosh, unpublished work). Thus dissecting further details of the control of AS160 and TBC1D1 by insulin/IGF-1, and the respective roles of these proteins in glucose homoeostasis, should give new insight into the links between insulin signalling, obesity and other metabolic disorders. AS160 and TBC1D1 are both widely expressed, and our findings also highlight AS160 as a potential mediator of growth-factor-regulated vesicle trafficking beyond insulin regulation of GLUT4.
Online data
Acknowledgments
This work was supported by the U.K. Medical Research Council, Diabetes U.K. and the companies who support the DSTT (Division of Signal Transduction Therapy) at the University of Dundee (Dundee, Scotland, U.K.), namely AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co, Merck KGaA and Pfizer. D. G. H. was supported by the EXGENESIS Integrated Project (LSHM-CT-2004-005272) of the European Commission. We thank to Colin Bell and Claire Balfour for tissue culture support, and Greg Stewart and the DSTT antibody production team co-ordinated by Dr James Hastie for affinity purification of antibodies. We thank Rachel Naismith for secretarial assistance and Kei Sakamoto for helpful discussions.
References
- 1.MacKintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benzinger A., Muster N., Koch H. B., Yates J. R., 3rd, Hermeking H. Targeted proteomic analysis of 14-3-3σ, a p53 effector commonly silenced in cancer. Mol. Cell. Proteomics. 2005;4:785–795. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 4.Meek S. E., Lane W. S., Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 5.Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harthill J. E., Pozuelo Rubio M., Milne F. C., MacKintosh C. Regulation of the 14-3-3-binding protein p39 by growth factors and nutrients in rat PC12 pheochromocytoma cells. Biochem. J. 2002;368:565–572. doi: 10.1042/BJ20020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozuelo Rubio M., Peggie M., Wong B. H., Morrice N., MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai S. L., Tee A. R., Short J. D., Bergeron J. M., Kim J., Shen J., Guo R., Johnson C. L., Kiguchi K., Walker C. L. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., Inoki K., Yeung R., Guan K. L. Regulation of TSC2 by 14-3-3 binding. J. Biol. Chem. 2002;277:44593–44596. doi: 10.1074/jbc.C200510200. [DOI] [PubMed] [Google Scholar]
- 10.Sekimoto T., Fukumoto M., Yoneda Y. 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27Kip1. EMBO J. 2004;23:1934–1942. doi: 10.1038/sj.emboj.7600198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem. J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howlett K. F., Sakamoto K., Garnham A., Cameron-Smith D., Hargreaves M. Resistance exercise and insulin regulate AS160 and interaction with 14-3-3 in human skeletal muscle. Diabetes. 2007;56:1608–1614. doi: 10.2337/db06-1398. [DOI] [PubMed] [Google Scholar]
- 13.Ramm G., Larance M., Guilhaus M., James D. E. A role for 14-3-3 in insulin-stimulated GLUT4 translocation through its interaction with the RabGAP AS160. J. Biol. Chem. 2006;281:29174–29180. doi: 10.1074/jbc.M603274200. [DOI] [PubMed] [Google Scholar]
- 14.Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 15.Perseghin G., Petersen K., Shulman G. I. Cellular mechanism of insulin resistance: potential links with inflammation. Int. J. Obes. Relat. Metab. Disord. 2003;27(Suppl. 3):S6–S11. doi: 10.1038/sj.ijo.0802491. [DOI] [PubMed] [Google Scholar]
- 16.Ishikura S., Bilan P. J., Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem. Biophys. Res. Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- 17.Larance M., Ramm G., Stockli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- 18.Miinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peranen J., Lane W. S., Lienhard G. E. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson H. K., Zierath J. R., Kane S., Krook A., Lienhard G. E., Wallberg-Henriksson H. Insulin-stimulated phosphorylation of the Akt substrate AS160 is impaired in skeletal muscle of type 2 diabetic subjects. Diabetes. 2005;54:1692–1697. doi: 10.2337/diabetes.54.6.1692. [DOI] [PubMed] [Google Scholar]
- 21.Watson R. T., Pessin J. E. Bridging the GAP between insulin signaling and GLUT4 translocation. Trends Biochem. Sci. 2006;31:215–222. doi: 10.1016/j.tibs.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Sano H., Kane S., Sano E., Miinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 23.Peck G. R., Ye S., Pham V., Fernando R. N., Macaulay S. L., Chai S. Y., Albiston A. L. Interaction of the Akt substrate, AS160, with the glucose transporter 4 vesicle marker protein, insulin-regulated aminopeptidase. Mol. Endocrinol. 2006;20:2576–2583. doi: 10.1210/me.2005-0476. [DOI] [PubMed] [Google Scholar]
- 24.Musi N., Hirshman M. F., Nygren J., Svanfeldt M., Bavenholm P., Rooyackers O., Zhou G., Williamson J. M., Ljunqvist O., Efendic S., et al. Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes. 2002;51:2074–2081. doi: 10.2337/diabetes.51.7.2074. [DOI] [PubMed] [Google Scholar]
- 25.Shaw R. J., Lamia K. A., Vasquez D., Koo S. H., Bardeesy N., Depinho R. A., Montminy M., Cantley L. C. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer H. F., Witczak C. A., Fujii N., Jessen N., Taylor E. B., Arnolds D. E., Sakamoto K., Hirshman M. F., Goodyear L. J. Distinct signals regulate AS160 phosphorylation in response to insulin, AICAR, and contraction in mouse skeletal muscle. Diabetes. 2006;55:2067–2076. doi: 10.2337/db06-0150. [DOI] [PubMed] [Google Scholar]
- 28.Thong F. S., Bilan P. J., Klip A. The Rab GTPase-activating protein AS160 integrates Akt, protein kinase C, and AMP-activated protein kinase signals regulating GLUT4 traffic. Diabetes. 2007;56:414–423. doi: 10.2337/db06-0900. [DOI] [PubMed] [Google Scholar]
- 29.Bruss M. D., Arias E. B., Lienhard G. E., Cartee G. D. Increased phosphorylation of Akt substrate of 160 kDa (AS160) in rat skeletal muscle in response to insulin or contractile activity. Diabetes. 2005;54:41–50. doi: 10.2337/diabetes.54.1.41. [DOI] [PubMed] [Google Scholar]
- 30.Treebak J. T., Glund S., Deshmukh A., Klein D. K., Long Y. C., Jensen T. E., Jorgensen S. B., Viollet B., Andersson L., Neumann D., et al. AMPK-mediated AS160 phosphorylation in skeletal muscle is dependent on AMPK catalytic and regulatory subunits. Diabetes. 2006;55:2051–2058. doi: 10.2337/db06-0175. [DOI] [PubMed] [Google Scholar]
- 31.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 32.Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. Phosphorylation of the protein kinase mutated in Peutz–Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90(RSK) and cAMP-dependent protein kinase, but not its farnesylation at Cys433, is essential for LKB1 to suppress cell growth. J. Biol. Chem. 2001;276:19469–19482. doi: 10.1074/jbc.M009953200. [DOI] [PubMed] [Google Scholar]
- 33.Williamson B. L., Marchese J., Morrice N. A. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol. Cell. Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Sapkota G. P., Cummings L., Newell F. S., Armstrong C., Bain J., Frodin M., Grauert M., Hoffmann M., Schnapp G., Steegmaier M., Cohen P., Alessi D. R. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. Inhibition of protein kinase Cμ by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 36.Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh M., Pullen N., Brennan P., Cantrell D., Dennis P. B., Thomas G. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J. Biol. Chem. 2002;277:20104–20112. doi: 10.1074/jbc.M201745200. [DOI] [PubMed] [Google Scholar]
- 38.Schalm S. S., Blenis J. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 39.Hardie D. G., Hawley S. A., Scott J. W. AMP-activated protein kinase: development of the energy sensor concept. J. Physiol. 2006;574:7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsin A. S., Bertrand L., Rider M. H., Deprez J., Beauloye C., Vincent M. F., Van den Berghe G., Carling D., Hue L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr. Biol. 2000;10:1247–1255. doi: 10.1016/s0960-9822(00)00742-9. [DOI] [PubMed] [Google Scholar]
- 41.Nakano M., Hamada T., Hayashi T., Yonemitsu S., Miyamoto L., Toyoda T., Tanaka S., Masuzaki H., Ebihara K., Ogawa Y., et al. α2 Isoform-specific activation of 5′adenosine monophosphate-activated protein kinase by 5-aminoimidazole-4-carboxamide-1-β-D-ribonucleoside at a physiological level activates glucose transport and increases glucose transporter 4 in mouse skeletal muscle. Metabolism. 2006;55:300–308. doi: 10.1016/j.metabol.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Karlsson H. K., Hallsten K., Bjornholm M., Tsuchida H., Chibalin A. V., Virtanen K. A., Heinonen O. J., Lonnqvist F., Nuutila P., Zierath J. R. Effects of metformin and rosiglitazone treatment on insulin signaling and glucose uptake in patients with newly diagnosed type 2 diabetes: a randomized controlled study. Diabetes. 2005;54:1459–1467. doi: 10.2337/diabetes.54.5.1459. [DOI] [PubMed] [Google Scholar]
- 43.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarbassov dos D., Ali S. M., Sabatini D. M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Corradetti M. N., Inoki K., Bardeesy N., DePinho R. A., Guan K. L. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz–Jeghers syndrome. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoki K., Zhu T., Guan K. L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 47.Shaw R. J., Bardeesy N., Manning B. D., Lopez L., Kosmatka M., DePinho R. A., Cantley L. C. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Stone S., Abkevich V., Russell D. L., Riley R., Timms K., Tran T., Trem D., Frank D., Jammulapati S., Neff C. D., et al. TBC1D1 is a candidate for a severe obesity gene and evidence for a gene/gene interaction in obesity predisposition. Hum. Mol. Genet. 2006;15:2709–2720. doi: 10.1093/hmg/ddl204. [DOI] [PubMed] [Google Scholar]
- 49.Roach W. G., Chavez J. A., Miinea C. P., Lienhard G. E. Substrate specificity and effect on GLUT4 translocation of the Rab GTPase-activating protein TBC1D1. Biochem. J. 2007;403:353–358. doi: 10.1042/BJ20061798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.