Abstract
Inorganic poly P (polyphosphate) is an abundant component of acidocalcisomes of Trypanosoma brucei. In the present study we report the presence of a protein homologous with the yeast Vtc1p (vacuolar transporter chaperone 1) in T. brucei that is essential for poly P synthesis, acidocalcisome biogenesis and cytokinesis. Localization studies in a cell line expressing a TbVTC1 fused to GFP (green fluorescent protein) revealed its co-localization with the V-H+-PPase (vacuolar H+-pyrophosphatase), a marker for acidocalcisomes. Western blot analysis of acidocalcisome fractions and immunogold electron microscopy using polyclonal antibodies against a fragment of TbVTC1 confirmed the acidocalcisome localization. Ablation of TbVTC1 expression by RNA interference caused an abnormal morphology of acidocalcisomes, indicating that their biogenesis was disturbed, with a decreased pyrophosphate-driven H+ uptake and Ca2+ content, a significant decrease in the amount of poly P and a deficient response to hyposmotic stress. Ablation of TbVTC1 expression for longer periods produced marked gross morphological alterations compatible with a defect in cytokinesis, followed by cell death. Overexpression of the TbVTC1 gene caused mild alterations in growth rate, but had no perceptible effect on acidocalcisome morphology. We propose that the PPi-driven H+ pumping deficiency induced by ablation of TbVTC1 leads to alterations in the protonmotive force of acidocalcisomes, which results in deficient fusion or budding of the organelles, decreased H+ and Ca2+ content, and decreased synthesis of poly P. A decrease in the poly P content would lead to osmotic sensitivity and defects in cytokinesis.
Keywords: acidocalcisome, polyphosphate, protonmotive force, Trypanosoma brucei, vacuolar H+ pyrophosphatase, vacuolar transporter chaperone
Abbreviations: BiP, immunoglobulin heavy-chain-binding protein; DAPI, 4′,6-diamidino-2-phenylindole; ECL, enhanced chemiluminescence; FBS, fetal bovine serum; GITC, guanidine isothiocyanate; GFP, green fluorescent protein; HRP, horseradish peroxidase; Iso-Cl, isotonic chloride; Nrf1, negative regulator of Cdc 42; ORF, open reading frame; PBST, PBS containing 0.1% (v/v) Tween 20; poly P, polyphosphate; Pi, inorganic phosphate; PPi, pyrophosphate; PPK, poly P kinase; RNAi, RNA interference; RT, reverse transcriptase; TbVTC1, Trypanosoma brucei vacuolar transporter chaperone 1; TbVP1, Trypanosoma brucei vacuolar H+ translocating pyrophosphatase 1; TEM, transmission electron microscopy; V-H+-PPase, vacuolar H+ translocating pyrophosphatase; VTC, vacuolar transporter chaperone; V-H+-ATPase, vacuolar H+ translocating ATPase
INTRODUCTION
Poly P (polyphosphate) is a linear chain of Pi (inorganic phosphate) moieties linked by high-energy phosphoanhydride bonds widely distributed from bacteria to mammals [1]. High levels of poly P accumulate in acidic organelles known as acidocalcisomes. Acidocalcisomes also contain large amounts of PPi (pyrophosphate) and bivalent cations such as Ca2+, Mg2+ and Zn2+. These organelles were first described in the protozoan parasites Trypanosoma brucei [2] and Trypanosoma cruzi [3], but have since been identified in an evolutionary diverse array of organisms from bacteria to humans [4].
T. brucei is the causative agent of sleeping sickness or African trypanosomiasis. According to the World Health Organization, over 60 million people in sub-Saharan Africa are at risk of infection with an incidence of 300000-500000 cases per year resulting in 55000 deaths. African trypanosomiasis has been re-emerging since 1970, and chemotherapy remains unsatisfactory, especially for advanced cases [5].
The synthesis of poly P in bacteria is performed by the action of PPKs (poly P kinases). In contrast relatively little is known about the synthesis of this polymer in eukaryotes [1]. Two PPKs have been described in bacteria: PPK1, which catalyses the reversible transfer of Pi residues from ATP to poly P and from poly P to ADP [6]; and PPK2, which catalyses the synthesis of poly P from GTP or ATP [7]. A PPK1 homologue to the bacterial PPK1 is present in the slime mould Dictyostelium discoideum (termed DdPPK1), which also has another PPK (termed DdPPK2) that is a tetramer composed of three actin-related proteins [8]. Using DNA microarray methodology, Ogawa et al. [9] identified the PHM1–PHM4 genes in Saccharomyces cerevisiae, which encode proteins that are involved in poly P synthesis, as demonstrated by the lack of detectable poly P in phm3Δ and phm4Δ mutants or in phm1Δ/phm2Δ double mutants. These results led the authors to speculate that the protein products of these genes might be poly P synthases [9]. Since then many protein sequence homologues from several organisms have been listed in the genome databases as poly P synthases. The PHM1–PHM4 genes were independently identified by Cohen et al. [10] and named VTC1–4 (VTC1/PHM4, VTC2/PHM1, VTC3/PHM2 and VTC4/PHM3) for vacuolar transporter chaperone [10,11], and a homologue to VTC1 was also found in Schizosaccharomyces pombe and named NRF1 (for negative regulator of the Cdc42 GTPase) [12,13]. The VTC1-null mutants had vacuoles that were fusion-incompetent [14], whereas the NRF1 null mutants had a severe endocytic defect [13]. Overexpression of NRF1 in S. pombe was lethal [13].
To investigate whether poly P synthesis in T. brucei is linked to the expression of a PHM4/VTC1/NRF1 gene homologue, in the present study we cloned and sequenced this gene, investigated the localization of its protein product and studied the effects of its ablation by RNAi (RNA interference) technology or its overexpression on parasite poly P synthesis, physiology and growth.
MATERIALS AND METHODS
Culture methods
Procyclic forms cell line 29-13 (T7RNAP NEO TETR HYG) co-expressing T7 RNA polymerase and tetracyclin repressor was a gift from Dr George A.M. Cross (Rockefeller University, New York, NY, U.S.A.) and was grown in SDM-79 medium supplemented with 10% FBS (fetal bovine serum) at 27 °C in the presence of G418 (15 μg/ml) and hygromycin (50 μg/ml) to maintain the integrated genes for T7 RNA polymerase and tetracycline repressor respectively [15]. Procyclic forms of the ILTar strain were grown in SDM-79 medium supplemented with 10% FBS.
Chemicals and reagents
TRIzol® reagent, Taq polymerase, SuperScript PCR buffer, Superscript II RT (reverse transcriptase), pCR 2.1-TOPO cloning kit, Alexa Fluor®-conjugated secondary antibodies, LysoTracker and 1-[2-(5-carboxyoxazol-2-yl)-6-aminobenzorufan-5-oxyl]-2-(2′-amino-5′-methylphenoxyl)-ethane-N,N,N′,N′-tetra-acetic acid and acetoxymethyl ester (fura 2/AM) were purchased from Invitrogen. The expression vector pET32a, Escherichia coli strain BL21trxB(DE3) and His-Bind Quick 900 resin columns were from Novagen. HRP (horseradish peroxidase)- and gold-conjugated antibodies were from Jackson ImmunoResearch Laboratories. An antibody against BiP (immunoglobulin heavy-chain-binding protein) was a gift from Dr Jay Bangs (University of Wisconsin, Madison, WI, U.S.A.). Rabbit and mouse polyclonal antibodies against TbVP1 (T. brucei vacuolar H+ translocating pyrophosphatase 1) were a gift from Dr Norbert Bakalara (Ecole Nationale Supérieure de Chimie de Montpellier, Montpellier, France) [16]. The p2T7Ti vector was a gift from Dr John Donelson (University of Iowa, Iowa City, IA, U.S.A.) [17]. The ECL® (enhanced chemiluminescence) detection kit was from Amersham Biosciences (GE Healthcare Life Sciences). The protein assay kit was from Bio-Rad. Primers were purchased from Integrated DNA Technologies. A monoclonal antibody against α-tubulin from sea urchin axonemes (clone B-5-1-2) and all other reagents of analytical grade were from Sigma.
Acidocalcisome fractions
Wild-type procyclic forms (ILTar strain) were collected and washed with cold PBS twice and then resuspended in lysis buffer [125 mM sucrose, 50 mM KCl, 4 mM MgCl2, 0.5 mM EDTA, 20 mM Hepes, 5 mM dithiotreitol and 1/500 protease inhibitor mixture Set III from Sigma (pH 7.2)]. Cell lysis and isolation of the acidocalcisome fraction using iodixanol gradient centrifugation was performed as described previously [18].
Generation of a TbVTC1–GFP (green fluorescent protein) fusion construct
To investigate the subcellular localization of TbVTC1 in parasites, the TbVTC1–GFP gene fusion was used, where Aequorea victoria GFP acts as a signal reporter gene. The T. brucei expression vector pUB39 with an inducible T7 promoter, provided by Dr George A. M. Cross, was used in the present study [15]. The full-length ORF (open reading frame) of TbVTC1 was amplified by RT-PCR using primers 5′-GAAGCTTATGGCAAGCGTAATTGAAAAG-3′ (a HindIII site is underlined) and 5′-GCTCGAGAGAAAATCGTGGTCCATCATT-3′ (an XhoI site is underlined); GFP gene was amplified using primers 5′-CCTCGAGGTGAGCAAGGGCGAGGAG-3′ (an XhoI site is underlined) and 5′-GGGATCCTTACTTGTACAGCTCGTCCAG-3′ (a BamHI site is underlined; a stop codon is indicated in bold). The resultant two PCR products were cloned into pCR 2.1-TOPO vectors respectively. Subsequently the TbVTC1 ORF was excised with HindIII and XhoI, and the GFP gene was excised with XhoI and BamHI. The two digested fragments were co-ligated into the linearized pUB39 vector, in which the inserted VSG117 cDNA fragment was removed by HindIII and BamHI digestion, and an in-frame fusion construct TbVTC1–GFP/pUB39 (Figure 1B, upper panel) was generated.
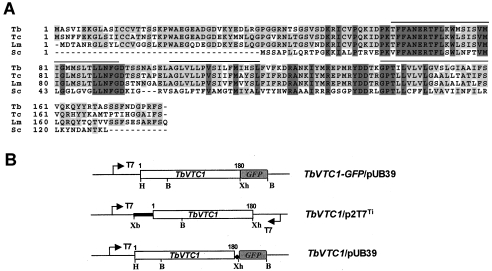
Figure 1. Sequence alignment of TbVTC1.
(A) ClustalW sequence alignment of TbVTC1 (Tb, AAX70699) with S. cerevisiae Phm4/Vtc1 (Sc, P40046) and two orthologues from T. cruzi (Tc, AY304574) and L. major (Lm, CAB 86965). Lines above the sequence represent predicted transmembrane domains. Identical residues are shaded. (B) Scheme of constructs used for GFP localization (upper), RNAi (middle) and overexpression (bottom) experiments. The arrows show the T7 promoters; the ◆ shows the introduced stop codon between TbVTC1 and GFP and the open boxes show the ORFs of TbVTC1 and GFP respectively. The bold line represents the 5′-untranslated region of TbVTC1. B, BamHI; H, HindIII; Xb, XbaI; Xh, XhoI.
Generation of TbVTC1 RNAi constructs
To ablate TbVTC1 mRNA in parasites by double-stranded RNA expression, the inducible T7 RNA polymerase-based protein expression system [15] and p2T7Ti vector with dual-inducible T7 promoters [17] were employed in the present study. A 654 bp DNA fragment (nucleotides −111 to 543) including the 5′-untranslated region and the entire ORF of TbVTC1 (Figures 1A and 1B, middle panel) was PCR-amplified using the forward primer 5′-GTCTAGATCCCCTTTGAGCGACCAACT-3′ and the reverse primer 5′-GCTCGAGAGAAAATCGTGGTCCATCATT-3′, where the underlined nucleotides indicate the introduced XbaI and XhoI sites respectively, allowing the TbVTC1 fragment to be cloned into the p2T7Ti vector (Figure 1B, middle panel).
Generation of a TbVTC1 overexpression construct
To study TbVTC1 overexpression in vivo, an additional recombinant construct was generated with a similar method as for the TbVTC1–GFP/pUB39 construct with a slight modification. Since the TbVTC1 gene contains one BamHI site (nucleotides 136–141; Figure 1B), and the pUB39 expression vector has only two restriction sites (HindIII and BamHI) available for foreign gene insertion, the BamHI site could not be used for TbVTC1 gene cloning into the pUB39 vector. A simple strategy was to introduce a stop codon (TGA) between the TbVTC1 and GFP genes in the TbVTC1–GFP/pUB39 construct, and the resultant construct allowed the expression of the TbVTC1 gene alone (Figure 1B, bottom panel). Therefore a modified reverse primer of TbVTC1 (5′-GCTCGAGTCAAGAAAATCGTGGTCCATCATT-3′, with an XhoI site underlined and a stop codon indicated in bold) was used.
All constructs were confirmed by DNA sequencing performed with the BigDye™ Terminator V3.0 Cycle Sequencing kit and a 373A DNA Automatic Sequencer (PerkinElmer Applied Biosystems).
Cell transfections
Exponential-phase procyclic forms (∼5×106 cells/ml) were harvested by centrifugation at 1500 g for 10 min, washed with Cytomix buffer [17] and resuspended in 0.45 ml of Cytomix buffer at a cell density of 2.5×107 cells/ml. The washed cells were mixed with 0.1 ml of NotI-linearized plasmid (5–10 μg) in a 0.4-cm electroporation cuvette and subjected to two pulses from a Bio-Rad Gene Pulser electroporator set at 1.5 kV and 25 μF. The stable transformants were selected in SDM-79 medium supplemented with 10% FBS plus 5 μg/ml phleomycin, 50 μg/ml hygromycin and 15 μg/ml G418. For induction of double-stranded RNA, 1 μg/ml fresh tetracycline was added to the cultures.
Antibody generation and purification
Expression of the full-length TbVTC1 in E. coli was unsuccessful for unknown reasons (results not shown). Therefore a truncated TbVTC1 fragment (coding the first 70 amino acids of TbVTC1) was amplified using the forward primer 5′-GGAATTCATGGCAAGCGTAATTGAA-3′ (an EcoRI site is underlined) and the reverse primer 5′-GCTCGAGTCAGGTCCTTTCATTGGCGA-3′ (an XhoI site is underlined; a stop codon is indicated in bold), and cloned into the expression vector pET32a. The resulting recombinant construct TbVTC1(1–70)/pET32a was transformed into E. coli BL21trxB(DE3) and the transformants were inoculated into Luria–Bertani broth supplemented with 50 μg/ml ampicillin and 30 μg/ml kanamycin at 37 °C. When the cultures reached a D600 of 0.5, expression was induced by the addition of 1 mM isopropyl β-D-thiogalactopyranoside. The induced cells were harvested after 4 h of incubation at 37 °C, washed with cold PBS and the pellets were kept frozen at −80 °C until use.
To purify the recombinant protein, the cell pellets were resuspended in binding buffer [300 mM NaCl, 50 mM Tris/HCl and 5 mM imidazole (pH 7.9)] and sonicated three times for 15 s with a Bronson Sonifier 450 at 15% amplitude with 30 s intervals on ice. After centrifugation at 20000 g for 30 min, the supernatant was loaded on pre-wetted His-Bind Quick 900 resin columns. Washing and elution steps were performed according to the manufacturer's protocol. The eluted fractions were pooled and dialysed against PBS at 4 °C overnight and concentrated with Microcon centrifugal filter devices. The purity of proteins was determined by SDS/PAGE.
A polyclonal antibody against recombinant TbVTC1 protein was generated in guinea-pigs by Cocalico Biologicals according to standard protocols. The antiserum was affinity-purified with the cyanogen-bromide-activated resin using standard protocols [19].
Fluorescence microscopy
T. brucei procyclic forms were harvested by centrifugation (1700 g, 10 min, 25 °C), washed with ice-cold PBS and fixed with 4% formaldehyde in PBS for 1 h at 4 °C. After washing with PBS, parasites were allowed to adhere to poly-L-lysine-coated coverslips, permeabilized with 0.3% Triton X-100 for 3 min, and blocked with PBS containing 3% BSA, 1% fish gelatin, 50 mM NH4Cl and 5% goat serum for 1 h. Cells were stained with the polyclonal rabbit anti-TbVP1 antibody (1:300 dilution) or a rabbit anti-BiP antibody (1:400 dilution) for 1 h. Cells were thoroughly washed with PBS and incubated with Alexa Fluor® 546-conjugated goat anti-rabbit antibodies at a dilution of 1:1000 for 45 min. The cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) before mounting with Gold ProLong Gold antifade reagent (Molecular Probes). The confocal images were collected with a Leica TCS SP2 laser-scanning confocal microscope.
For labelling with LysoTracker, fresh cells were washed with SDM-79 medium (pre-warmed to 27 °C) twice, and then resuspended at a concentration of 1×107 ml−1 in SDM-79 medium with 100 nM LysoTracker for 30 min. The stained cells were washed, fixed and adhered to coverslips. DIC (differential interference contrast) and fluorescent optical images were immediately captured under non-saturating conditions and identical exposure times using an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ CCD (charge-coupled device) camera driven by DeltaVision software (Applied Precision).
Western blot analysis
The procyclic forms were harvested, washed twice in PBS and resuspended in PBS containing proteinase inhibitors (1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1 mM PMSF). The cells were broken with five cycles of freezing and thawing. The total cell lysates were mixed with 2× Laemmli sample buffer, boiled for 5 min, and analysed by SDS/PAGE (15% gels). The separated proteins were transferred on to nitrocellulose membranes (Osmonics) using a Bio-Rad transblot apparatus. The membranes were blocked with 5% non-fat dried skimmed milk in PBST [PBS containing 0.1% (v/v) Tween 20] for 2 h at room temperature (25 °C). The blots were incubated with the purified guinea-pig anti-TbVTC1 polyclonal antibody at a dilution of 1:3000 or an anti-α-tubulin monoclonal antibody at a dilution of 1:1500 for 1 h. After five washings with PBST, the blots were incubated with HRP-conjugated goat anti-guinea-pig or goat anti-mouse IgG [H +L (heavy +light chain)] antibody at a dilution of 1:20000 for 1 h. After washing five times with PBST, the immunoblots were visualized using the ECL® detection kit according to the manufacturer's instructions.
Electron microscopy
Approx. 1×108 procyclic forms were harvested and washed twice with cold PBS. The parasites were fixed with freshly prepared 2.5% glutaraldehyde, 4% paraformaldehyde and 0.1 M sodium cacodylate buffer (pH 7.3) on ice for 1 h and then embedded in epoxy resin, sectioned and stained using standard methods. Immunogold electron microscopy experiments were performed as described previously [20] with a purified anti-TbVTC1 antibody (1:200) and mouse polyclonal antibody against TbVP1 (1:10). After washing, the grids were incubated with 18 nm colloidal gold-AffiniPure conjugate donkey anti-guinea-pig IgG (H+L) and 12 nm colloidal gold conjugate goat anti-mouse IgG (H+L). Images were acquired on a Phillips CM-200 transmission electron microscope operating at 120 kV. For scanning electron microscopy, cells were fixed as before, adhered to poly-L-lysine-coated grids, dehydrated in ethanol series, critical point dried and coated with gold in a Balzers gold sputtering system. Cells were observed in a Leo environmental scanning electron microscope operating at 15 kV. For imaging whole procyclic forms, cells were washed with filtered buffer A [116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 50 mM Hepes (pH 7.2) and 5.5 mM glucose] twice, and directly applied to Formvar-coated copper grids, allowed to adhere for 10 min, carefully blotted dry, and observed in an energy-filtering Zeiss EM 902 electron microscope operating at 80 kV. Electron spectroscopic images were recorded at an energy loss of 60 eV using a spectrometer slit width of 20 eV. For determination of morphometric parameters such as number, circularity, diameter and absolute volume of acidocalcisomes in wild-type cells and cells in which knockdown of TbVTC1 was performed, whole unfixed parasites were observed and randomly selected in a Zeiss EM 902 transmission electron microscope equipped with an energy filter. For estimation of the absolute volume of acidocalcisomes, the diameters of 50 organelles that were not overlapping were measured; their volumes were then calculated on the assumption that they were spherical units. Statistical significance was determined by Student's t test. P<0.05 was taken to be significant.
Calcium analysis and H+ pump activity
Calcium stores were assessed after loading cells with fura 2 as previously described [3]. Acidification of internal compartments was followed by measuring changes in the absorbance of Acridine Orange at the wavelength pair 493–530 nm in an Olis-modified SLM-Aminco DW2000 dual-wavelength spectrophotometer [21]. Procyclic forms were incubated at 30 °C in 2.5 ml standard reaction medium containing 125 mM sucrose, 65 mM KCl, 2 mM MgCl2, 10 mM Hepes buffer (pH 7.4), with or without 1.5 μM digitonin before addition of 3 μM Acridine Orange. The results shown are representative of at least three experiments.
Extraction of long-chain and short-chain poly P and poly P measurements
Cells (1×107) were harvested and washed with buffer A [116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 50 mM Hepes and 5.5 mM glucose (pH 7.2)] twice. For short-chain poly P extraction, the cell pellet was resuspended in ice-cold 0.5 M perchloric acid (HClO4) and incubated on ice for 30 min. After centrifugation for 3000 g for 5 min, the supernatant was neutralized with the addition of 0.72 M KOH/0.6 M KHCO3. The precipitated KClO4 was removed by centrifugation at 12000 g for 10 min and the supernatant was transferred to a new tube for poly P determination. The long-chain poly P was extracted with glassmilk as described by Ault-Riché et al. [22]. Briefly, the cell pellet was resuspended with 500 μl of GITC-lysis buffer [4 M guanidine isothiocyanate and 50 mM Tris/HCl (pH 7.0)] pre-warmed at 95 °C with vortex-mixing for several seconds. The cell mixture was incubated in a heat block at 95 °C for 2–5 min and sonicated briefly. To each tube 30 μl of 10% SDS, 500 μl of ethanol and 5 μl of glassmilk (Qbiogene) were added. After incubating for 5 min with brief vortex-mixing, the tube was centrifuged at 14000 g for 20 s and the pelleted glassmilk was washed with 0.5 ml of cold freshly prepared washing buffer [5 mM Tris/HCl, 50 mM NaCl, 5 mM EDTA and 50% (v/v) ethanol (pH 7.5)] three times. The washed pellet was resuspended with 50 μl of 50 mM Tris/HCl and 10 mM MgCl2 (pH 7.5) containing 20 μg/ml DNase and 20 μg/ml RNase. After incubation in a water bath at 37 °C for 10 min, the pellet was washed once with 150 μl of GITC-lysis buffer and 150 μl of ethanol, and then twice with washing buffer. Finally poly P was eluted with 50 μl of 50 mM Tris/HCl (pH 8.0) at 95 °C for 2 min and collected by centrifugation at 14000 g for 20 s. The extracted poly P was immediately measured as described below. The remaining poly P was stored at −80 °C.
For poly P measurements, the recombinant exopolyphosphatase of S. cerevisiae (rScPPX1) was expressed and purified by affinity chromatography from E. coli strain CA38pTrcPPX1 (a gift from Dr Arthur Kornberg). The purity of rScPPX1 was determined by SDS/PAGE. Poly P levels were determined by the amount of Pi released upon treatment with an excess of rScPPX1. The enzymatic reaction was performed in 96-well plates with 50 mM Tris/HCl (pH 7.4), 6 mM MgCl2, 3000–5000 units of purified rScPPX1 and 5 μl of extracted poly P sample at a final volume of 100 μl (1 unit corresponds to the release of 1 pmol of Pi/min at 37 °C). After incubation at 37 °C for 15 min, the reaction was immediately stopped by the addition of an equal amount of the fresh mixture of three parts of 0.045% Malachite Green with one part of 4.2% ammonium molybdate (Sigma), which was filtered prior to use as described previously [23]. The absorbance at 660 nm was read using a SpectraMax M2e plate reader (Molecular Devices) after incubating for 10 min at room temperature for colour development. The free Pi (released) amount was determined by using a standard curve of pmol of phosphate against D660. The poly P concentration was calculated based on the intracellular cell volume of 0.48±0.02 μl per 107 T. brucei procyclic form cells [24].
Regulatory volume decrease
The cells were collected and washed twice with isotonic chloride buffer [Iso-Cl buffer; 137 mM NaCl, 4 mM KCl, 1.5 mM KH2PO4, 8.5 mM Na2HPO4, 20 mM Hepes, 11 mM glucose, 1 mM CaCl2 and 0.8 mM MgSO4 (pH 7.4)]. The buffer osmolarity was 300±5 mOsm (‘isosmotic’) as verified by an Advanced Instruments 3D3 osmometer. The washed cells were resuspended in Iso-Cl buffer to a cell density of 1×108/ml for wild-type cells and 0.5×108/ml for TbVTC1 mutant cells (equivalent amounts of mg of protein, since most mutant cells were fat and enlarged) respectively. The cells were distributed in 96-well plates with 150 μl per well in triplicate and 150 μl of sterile deionized water was added to each well using a multichannel pipettor, which resulted in a final osmolarity of 150 mOsm at time zero. The absorbance changes were followed at 550 nm for 10 min using an SpectraMax M2e plate reader to monitor the cell volume changes as described previously [25].
RESULTS
Sequence analysis of TbVTC1
We identified a Phm4p/Vtc1p homologue in the T. brucei genome by searching against the database with the S. cerevisiae Phm4p/Vtc1p sequence. The TbVTC1 gene encodes a protein of 180 amino acid residues with a predicted molecular mass of 19.8 kDa and a calculated isoelectric point of 8.85. Topology modelling predicted three transmembrane domains (residues 70–92, 102–121 and 142–161 respectively), an intracellular N-terminus and an extracellular C-terminus (results not shown). Similar sequences were also found in the Leishmania major and Trypanosoma cruzi databases. (Figure 1A) shows a sequence alignment of the S. cerevisiae PHM4/VTC1 protein with the deduced amino acid sequences of the T. brucei, L. major and T. cruzi gene products. According to this alignment, the T. brucei gene product was 75, 71 and 33% identical with the T. cruzi, L. major and S. cerevisiae gene products respectively.
Subcellular localization of TbVTC1
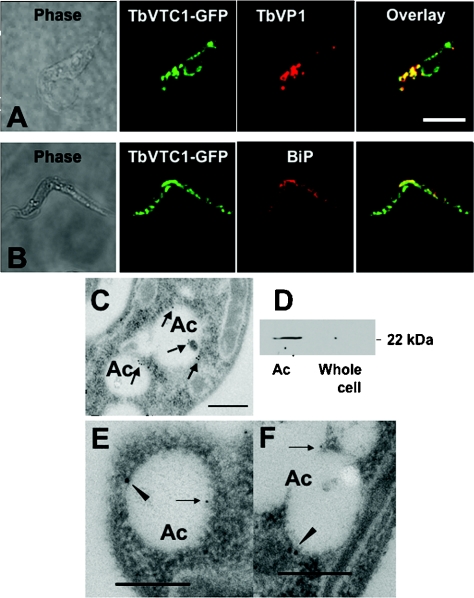
To investigate the localization of TbVTC1, a TbVTC1–GFP fusion expression construct was generated (Figure 1B, top panel) and transfected into procyclic forms. After induction with a low concentration of tetracycline (0.5 μg/ml) and for a short period (2 h) to avoid mistargeting, transformants expressed TbVTC1–GFP, which had punctate and perinuclear localization. The punctate staining co-localized with the reaction of antibodies against TbVP1 (Figure 2A), a marker of acidocalcisomes [26], whereas the perinuclear localization corresponded to the endoplasmic reticulum, as indicated by its co-localization with an antibody to the endoplasmic reticulum marker BiP (Figure 2B). The latter co-localization was most probably due to protein overexpression. It is interesting to note, however, that Vtc1p localizes in the vacuole and also in the endoplasmic reticulum of S. cerevisiae [27].
Figure 2. TbVTC1 localizes in acidocalcisomes and endoplasmic reticulum.
TbVTC1–GFP partially co-localized with the TbVP1 in acidocalcisomes (A) and with BiP in the endoplasmic reticulum (B) as shown by immunofluorescence microscopy analysis. Scale bars: 10 μm. Localization in acidocalcisomes (Ac) was confirmed by immunogold labelling with an anti-TbVTC1 antibody in wild-type cells through electron microscopy analysis (C). TbVTC1 (closed arrowheads, 18 nm gold) also co-localized with TbVP1 (arrows, 12 nm gold) in acidocalcisomes (E and F). Scale bars: 0.2 μm. After subcellular fractionation of wild-type cells, TbVTC1 was preferentially detected by Western blot analysis in the acidocalcisome fraction (Ac) with no detection with an equal amount of protein (15 μg), in a whole-cell lysate (D).
Although purified antibody raised against TbVTC1 performed well in Western blot analysis (Figure 2D, and see below), it did not produce signal when used in immunofluorescence experiments. Therefore the results of the TbVTC1–GFP localization could not be confirmed by immunofluorescence with an anti-TbVTC1 antibody. However, affinity-purified antibody did produce satisfactory signals by immunogold electron microscopy in wild-type (ILTar strain) procyclic forms (Figures 2C, 2E and 2F), which served to confirm that the GFP results were not due to mistargeting. (Figures 2E) and (2F) show that TbVTC1 (arrowheads) co-localized with TbVP1 (arrows), the acidocalcisome marker.
To further confirm the subcellular location of TbVTC1, we performed experiments on isolated acidocalcisomes. A Western blot analysis with an anti-TbVTC1 antibody showed a strong band at 22 kDa in the acidocalcisome fraction, which was close to the predicted mass of the protein, whereas an equal amount of protein of a whole-cell extract (15 μg) showed no signal (Figure 2D), consistent with a significant concentration of the protein in the acidocalcisome.
Effect of RNAi ablation or overexpression of TbVTC1 on cell growth and morphology
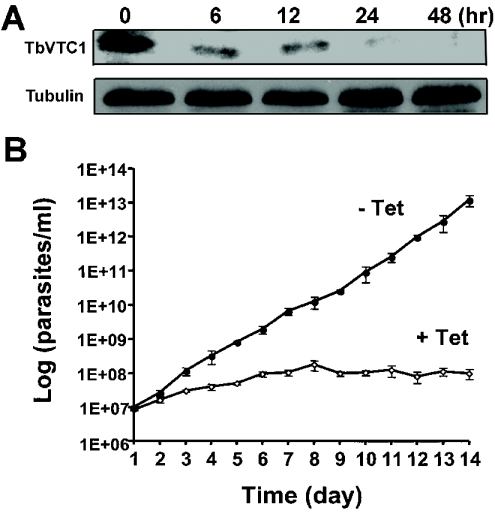
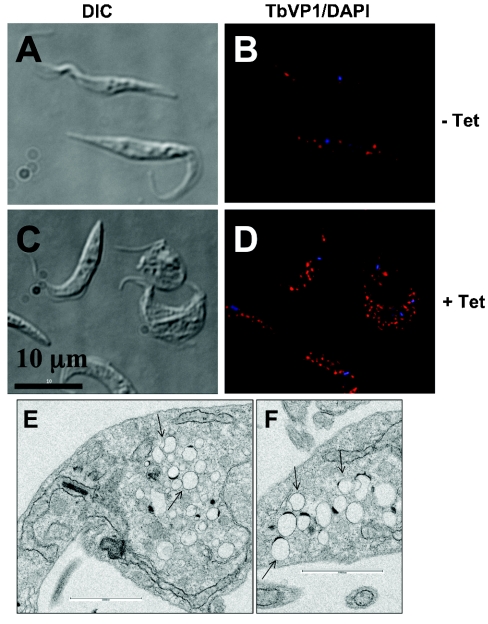
After transfection of the procyclic forms with the RNAi vector construct as detailed in the Materials and methods section, TbVTC1 ablation was induced by addition of tetracycline. Western blot analysis with an anti-TbVTC1 antibody showed that protein expression was down-regulated within 24 h of addition of 1 μg/ml tetracycline to an exponential-phase culture (Figure 3A). In addition, beginning on approx. day 3–4 after tetracycline induction, cell growth was almost completely inhibited (Figure 3B). This cessation of cell growth correlated with the appearance of gross morphological alterations. By day 4 numerous round, enlarged cells were observed (see Supplementary Figure 1A, right-hand panel, at http://www.BiochemJ.org/bj/407/bj4070161add.htm), and by 1 week the morphological disruptions had progressed to include, for example, anucleate ghosts containing only kinetoplast DNA (see Supplementary Figure 1A, left-hand panel), and multinucleate (see Supplementary Figure 1B, middle panel) or multiflagellate (see Supplementary Figure 1B, right-hand panel) cells. Scanning electron microscopy of the induced cells further revealed significant alterations in the size of the cells (see Supplementary Figures 2B, 2C and 2E at http://www.BiochemJ.org/bj/407/bj4070161add.htm), flagellar detachment from the surface of the cells (see Supplementary Figures 2B and 2D) and multiflagellate cells (see Supplementary Figure 2D). Non-induced control cultures grew normally (Figure 3B) and exhibited normal morphology (see Supplementary Figure 1A, left-hand panel, and Figure 2A).
Figure 3. Effect of RNAi of TbVTC1 on the growth and morphology of procyclic forms.
(A) At 24 h after addition of tetracycline, TbVTC1 expression could no longer be detected by Western blot analysis. Each lane was loaded with 40 μg of protein of whole cell lysates. Note that the amount of protein loaded was higher and the exposure time was longer than in (Figure 2D), to obtain valid comparisons with signals obtained after induction (upper panel). The blot was reprobed with monoclonal antibody against α-tubulin as a loading control (lower panel). (B) Effect of RNAi ablation on cell growth (□) compared with control growth without the addition of tetracycline (●). 1E+06 etc., 1 × 106 etc.
Procyclic forms transfected with the TbVTC1 overexpression vector construct as detailed in the Materials and methods section were induced to overexpress TbVTC1 by the addition of 1 μg/ml tetracycline. Western blot analysis with an anti-TbVTC1 antibody showed that the exogenous TbVTC1 expression increased significantly within 6 h of tetracycline addition (see Supplementary Figure 3A at http://www.BiochemJ.org/bj/407/bj4070161add.htm). TbVTC1 overexpressing cells showed a slight decrease in growth rate for the first 7 days of culture but then recovered (see Supplementary Figure 3B). Cell morphology was normal throughout the growth period (results not shown).
Effect of TbVTC1 RNAi ablation on acidocalcisome morphology and function
After 4 days of tetracycline induction, when the enlarged cell phenotype (see Supplementary Figure 1A, right-hand panel) was just becoming apparent, cells were fixed and stained with an antibody against TbVP1, or prepared as a whole mount for TEM (transmission electron microscopy). Non-induced cells showed a high number (36±11) of acidocalcisomes (as seen in TEM images, see below) located throughout the cell body. However, induced cells showed dramatic changes in acidocalcisome numbers. A certain percentage of cells possessed higher numbers of acidocalcisomes (Figure 4D), whereas a proportion of cells had fewer than normal amounts of these organelles (see below). TEM of thin sections confirmed that acidocalcisomes completely filled entire subcellular regions of some parasites, whereas mitochondrial morphology remained essentially normal (Figures 4E and 4F). Control experiments using up to 2.5 μg/ml tetracycline for at least 5 days did not produce any effect on wild-type procyclic forms (results not shown).
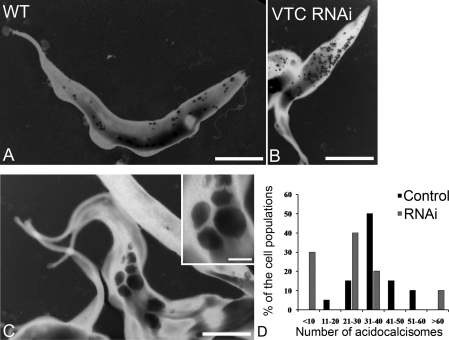
Figure 4. Morphology of acidocalcisomes after ablation of TbVTC1 by RNAi.
Acidocalcisome morphology was assessed before (A and B) and after (C and D) the addition of tetracycline for 4 days by fixing and staining the cells with antibody against TbVP1 (B and D; red). The DAPI staining showed the nuclei and kinetoplasts (blue). Scale bars in (A–D): 10 μm. In addition, the transmission electron microscopic analysis (E and F) after tetracycline induction showed large regions of the cells densely packed with small acidocalcisomes (arrows). Scale bars in (E) and (F): 0.2 μm.
To investigate the changes in acidocalcisome numbers in more detail we analysed whole cells by energy-filtering TEM (Figures 5A–5C), and estimated the average number of acidocalcisomes per cell in the induced and non-induced populations (Figure 5D). The number of acidocalcisomes per cell in non-induced cells followed a bell curve distribution. In contrast, when induced with tetracycline there were three major phenotypes in VTC1 RNAi cells: (i) cells with fewer than ten acidocalcisomes, representing 30% of the population; (ii) cells with a number of acidocalcisomes between 11 and 60, accounting for 60% of the population; and (iii) cells with a large number of acidocalcisomes (>60), which represented 10% of the population (Figure 5D). Nevertheless the average number of acidocalcisomes in both induced and non-induced cells was approx. 30, resulting in no statistical difference in the total number of these organelles in the two groups of cells. Those cells with lower numbers had bigger acidocalcisomes (Figure 5C), whereas those with higher numbers had smaller acidocalcisomes (Figure 5B), resulting in statistically significant differences in the total volume occupied by these organelles in wild-type cells and all cells subjected to RNAi (Table 1).
Figure 5. Morphological changes after RNAi of TbVTC1 and numeric distribution of acidocalcisomes in T. brucei.
(A–C) TEM of whole procyclic trypomastigotes. The dark granules are the acidocalcisomes. (A) Wild-type (WT) procyclic stages; (B and C) procyclic forms after 4 days of induction with tetracycline. (B) shows cells with numerous small acidocalcisomes, whereas (C) and inset show cells with large acidocalcisomes. Scale bars: (A) 3 μm, (B) 5 μm and (C) 4 μm. (D) Whole unfixed parasites were observed using a Zeiss EM 902 transmission electron microscope equipped with an energy filter and the number of acidocalcisomes per cell in ∼50 cells was counted.
Table 1. Morphometric analysis of the acidocalcisomes in T. brucei.
The number, circularity, diameter and absolute volume (calculated assuming acidocalcisomes are spheres) of the organelles were analysed and compared in control and TbVTC1 ablated cells. The cells submitted to RNAi (TbVTC1RNAi) were subdivided into three groups representing cells with fewer than ten, between 11 and 60, and more than 60 acidocalcisomes per cell. Results are expressed as means±S.D. *Significance as compared with wild-type cells (P<0.05).
| Number of acidocalcisomes | Mean circularity (nm) | Mean diameter (nm) | Absolute volume (×106 nm3) | |
|---|---|---|---|---|
| Wild-type | 36±11 | 0.95±0.02 | 194.6±19.26 | 4.83±1.69 |
| TbVTC1 RNAi | ||||
| ≤10 | 8±2* | 0.82±0.009* | 674.2±194.4* | 259.5±91.7* |
| 11–60 | 28±7 | 0.84±0.07* | 432.1±84.7* | 82.3±60.5* |
| ≥60 | 117±46* | 0.89±0.01* | 316.7±42.1* | 25.7±12.3* |
Decreased V-H+-PPase (vacuolar H+ translocating pyrophosphatase) activity after RNAi of TbVTC1
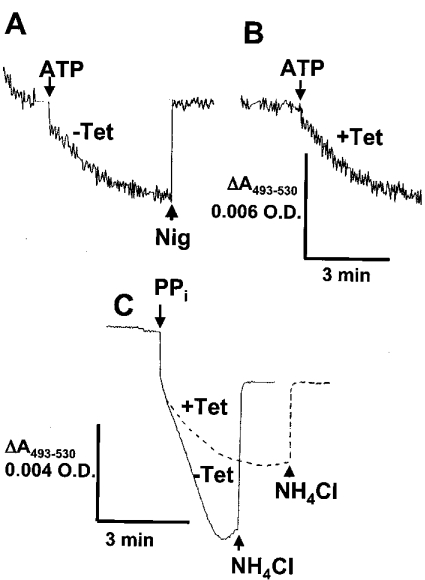
Since a defect in acidocalcisome fusion was apparent (Figures 5A–5C) and it has been shown in yeast that a protonmotive force across the membrane is required for vacuolar fusion [28–30], we tested whether acidocalcisomes of TbVTC1-knockdown cells were defective for H+ pumping. Previous studies demonstrated the feasibility of detecting acidocalcisomal H+ transport using cells permeabilized with digitonin [26,31]. Acidocalcisomes from T. brucei possess two H+ pumps, a V-H+-ATPase (vacuolar H+ translocating ATPase) [2] and the V-H+-PPase [26]. As reported previously [26], when procyclic trypomastigotes were permeabilized with digitonin, some Acridine Orange accumulated and was retained in the absence of energy sources (Figure 6A). Once a steady state of Acridine Orange accumulation was reached, addition of 1 mM ATP led to additional dye uptake (Figure 6A). The gradient collapsed completely after addition of 1 μM nigericin (Figure 6A). We have reported previously that this ATP-driven H+ uptake is completely inhibited by pre-incubation with 0.5 μM bafilomycin A1 [21]. Experiments with procyclic trypomastigotes in which TbVTC1 was ablated by RNAi revealed similar ATP-driven H+ uptake (Figure 6B). However, in parasites deficient in TbVTC1, the H+ translocation activity was reduced to 40% of that of wild-type cells when PPi was used as a substrate instead of ATP (Figure 6C), revealing a deficient V-H+-PPase activity. Addition of NH4Cl collapsed the H+ gradient generated by the V-H+-PPase (Figure 6C).
Figure 6. ATP- and PPi-driven H+ transport in permeabilized procyclic trypomastigotes.
Procyclic trypomastigotes [non-induced (A and C, continuous line) or after 4 days of induction with tetracycline (B and C, broken line); 0.1 mg of protein/ml] were added to a medium containing 125 mM sucrose, 65 mM KCl, 2 mM MgCl2, 10 mM Hepes (pH 7.4), 1.5 μM digitonin and 3 μM Acridine Orange. ATP (1 mM), PPi (0.2 mM), nigericin (1 μM) or NH4Cl (10 mM) was added where indicated. The scale for (A) and (B) is the same and is shown under (B). The results shown are representative of at least three experiments with different cell preparations.
Deficient H+ gradient and poly P synthesis after ablation of TbVTC1
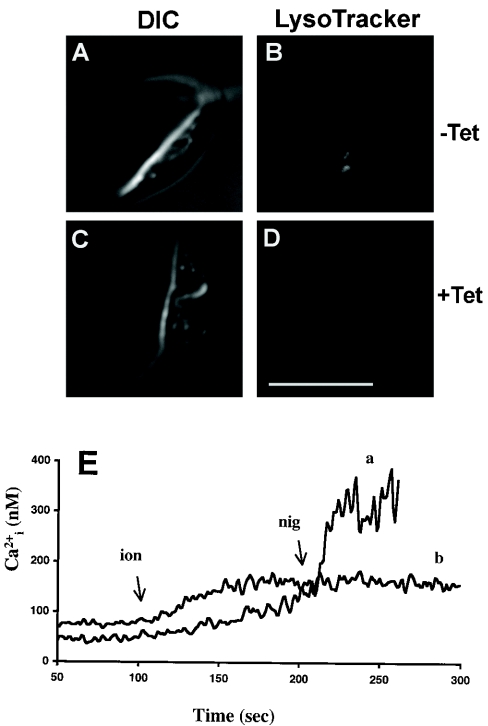
We have reported previously that the H+ gradient generated by the V-H+-PPase is necessary for the synthesis of poly P [31], as well as for the Ca2+ pumping activity of the acidocalcisomal Ca2+/H+-countertransporting ATPase [2,32]. We therefore examined the effects of TbVTC1 ablation on their H+, Ca2+ and poly P content. After 4 days of induction with tetracycline, the acidocalcisomes were noticeably less acidic, as assessed by their inability to accumulate the dye LysoTracker (Figure 7D), which specifically accumulates in acidic compartments. This was in contrast with non-induced cells, which accumulated the dye avidly (Figure 7B). Additionally, in cells loaded with the Ca2+-sensitive fluorophore fura 2, subcellular acidic Ca2+ stores, which can be specifically assessed by the sequential addition of the Ca2+ ionophore ionomycin and the K+/H+ exchanger nigericin [32], were greatly reduced (Figure 7E).
Figure 7. RNAi of TbVTC1 reduces H+ and Ca2+ content of acidocalcisomes.
(A–D) Control (A and B) and tetracycline-induced (C and D) procyclic forms (4 days of induction) were collected and washed twice with fresh SDM-79 medium plus 10% FBS. The cells were resuspended and stained with 100 nM LysoTracker. The control cells accumulated LysoTracker avidly (B). After ablation of TbVTC1 by RNAi, acidocalcisomes were alkalinized as evident by their inability to take up the acidophilic dye LysoTracker (D). Scale bars: 10 μm. (E) Acidic Ca2+ stores, assessed by sequential addition of 1 μM ionomycin and nigericin in fura 2-loaded cells, were also reduced in TbVTC1-ablated cells (E, trace b) compared with control cells (trace a).
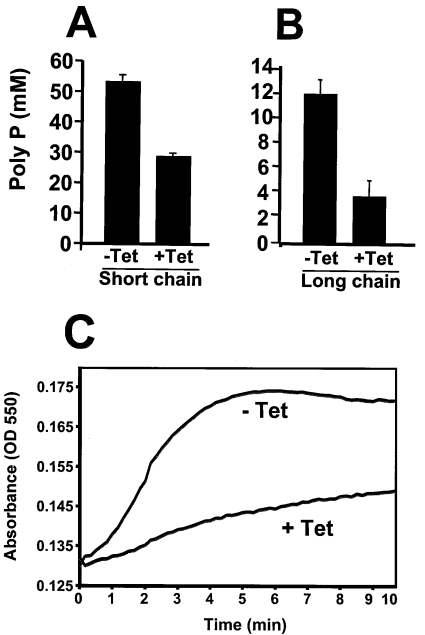
TbVTC1-ablated cells had significantly reduced levels of both short-chain (<50 phosphate units) and long-chain (>50 up to 700–800 phosphate units) [6] poly P compared with controls (Figures 8A and 8B). The decrease in long-chain poly P was more evident than that of short-chain poly P.
Figure 8. Levels of short-chain (A) and long-chain (B) poly P and regulatory volume decrease (C) in procyclic trypomastigotes before and after induction of TbVTC1 ablation.
(A and B) Poly P levels were determined as described in the Materials and methods section. (C) TbVTC RNAi cells (−Tet, non-induced; +Tet, induced for 4 days with tetracycline) suspended in Iso-Cl buffer were diluted with water to a final osmolarity of 150 mOsm at time zero, and relative changes in cell volume were followed by monitoring the absorbance at 550 nm as described in the Material and methods section. Note that the initial decrease in absorbance is too fast to be recorded.
Ablation of TbVTC reduces the ability of the cells to recover their volume after hyposmotic stress
A regulatory volume decrease in response to hyposmotic stress has been characterized in different trypanosomatids and a role for acidocalcisomes and poly P hydrolysis in this process has been documented [25,31]. When procyclic trypomastigotes were subjected to a 50% reduction in osmolarity (from 300 to 150 mOsm) they rapidly swelled but then began to shrink within a few seconds, such that after 5 min they were virtually indistinguishable in terms of motility and morphology from control cells maintained under isosmotic conditions (results not shown). These observations were confirmed by following volume recovery over time using the light-scattering technique described previously [25], in which changes in absorbance of a cell suspension are negatively correlated with changes in cell volume. Procyclic trypomastigotes in which TbVTC1 was ablated by RNAi were dramatically deficient in their regulatory volume decrease (Figure 8C).
DISCUSSION
In the present study we have identified and characterized a homologue to the S. cerevisiae Phm/Vtc family in T. brucei. In yeast these proteins have been implicated in several membrane-related processes, such as sorting of H+-ATPases, endocytosis, endoplasmic reticulum-Golgi trafficking, vacuole fusion, vacuolar poly P homoeostasis and microautophagy [9–14,27]. In the present study we have also demonstrated the role of TbVTC1 in acidocalcisome biogenesis and function.
In procyclic forms of T. brucei, a VTC1–GFP fusion construct localized to the acidocalcisomes (Figure 2A) and immunogold electron microscopy and subcellular fractionation studies (Figures 2C–2F) confirmed the localization of TbVTC1 in acidocalcisomes.
Ablation of TbVTC1 by RNAi led, within 4 days, to cessation of cell growth and to abnormal morphology of acidocalcisomes, indicating that their biogenesis was disturbed. These changes were accompanied by a deficient PPi-driven H+ pumping activity in permeabilized cells. Most changes observed in the cells could be explained by this H+ pumping deficiency. We have demonstrated previously that PPi-driven H+ pumping is able to generate a membrane potential in isolated acidocalcisomes [26] and that this protonmotive force is needed for their poly P synthesis [31]. This would explain the decrease in poly P content of the cells after RNAi (Figures 8A and 8B). Since the H+ gradient is needed for the function of the Ca2+/H+ countertransporting ATPase [2,26], this would also explain the decrease in Ca2+ content of acidocalcisomes (Figure 7E). In addition, we also demonstrated that acidocalcisomes and poly P are involved in the regulatory volume decrease mechanism that occurs as a consequence of hyposmotic stress [20], which would explain the inhibition of the regulatory volume decrease after RNAi of TbVTC1 (Figure 8C).
Acidocalcisomes are very dynamic and capable of budding or fusing with each other or with other organelles such as the contractile vacuole [4]. It has been shown that the protonmotive force is important for membrane fusion of vacuoles [28–30]. As a consequence of a diminished protonmotive force it is possible that acidocalcisomes of tetracycline-induced cells have defects in membrane fusion and that the phenotypes observed are due to acidocalcisomes that did not fuse (in the case of presence of numerous small organelles) and to acidocalcisomes that did not bud (in the case of few and giant acidocalcisomes).
After prolonged RNAi of TbVTC1 the appearance of bizarre cell morphologies are compatible with a defect in cytokinesis (see Supplementary Figures 1A and 1B at http://www.BiochemJ.org/bj/407/bj4070161add.htm). This included the development of multinucleate and multiflagellate cells by 1 week. It is interesting to speculate as to why ablation of a VTC1 protein localized in acidocalcisomes led to an apparent defect in cell division. One possibility is simply that this is a toxic phenotype secondary to the dramatic perturbations of Ca2+ and poly P homoeostasis. Dramatic toxic morphologies are common in RNAi experiments in T. brucei [33,34], and it can be difficult to tease out specific from non-specific effects. However, another possibility is afforded by the fact that a VTC1 homologue in S. pombe (Nrf1) is an important regulator of Cdc42p [12]. Cdc42p is a small Rho-like GTPase, which is found in most eukaryotic cells [35] and, in addition to being important for vacuolar function and morphology in yeast, is also critical for cell polarity and cytokinesis [36,37]. Most interestingly, Cdc42p is apparently critical for docking assembly in S. cerevisiae [38].
Finally, it should be noted that many Apicomplexan and trypanosomatid parasites have database sequences with homology with Phm/Vtc proteins (see Supplementary Figure 4 at http://www.BiochemJ.org/bj/407/bj4070161add.htm). A wider search of the database revealed a large number of putative proteins with homology with the yeast Phm/Vtc family. These included sequences from Toxoplasma, Plasmodium and Cryptosporidium. Previously, studies on yeast had explored the homology of the N-termini of the Phm1p/Vtc2, Phm2p/Vtc3 and Phm3p/Vtc4 (with molecular masses of 95.4, 96.6 and 75.5 kDa respectively). However, these studies neglected the prototypical Phm4p/Vtc1p because it is much smaller (14.4 kDa) than the other three yeast isoforms identified. Our database studies revealed that indeed there were both small and large proteins with Phm/Vtc homology. Large homologues (66.1–129.0 kDa) were detected in S. pombe, Candida albicans, Encephalitozoon cuniculi, Toxoplasma gondii, Cryptosporidium hominis, Cryptosporidium parvum, Plasmodium berghei, Plasmodium chabaudi, Plasmodium falciparum, L. major, T. brucei and T. cruzi. Short homologues (13.4–19.9 kDa) to S. cerevisiae Phm4p/Vtc1 were detected in S. pombe, T. cruzi, T. brucei and L. major. Regardless of size, all proteins examined shared a conserved motif, located centrally (T. brucei, T. cruzi and L. major) or near the N-terminus (S. cerevisiae and S. pombe) in the case of the short Phm4p/Vtc1 homologues and near the C-terminus in the case of the long homologues (see Supplementary Figure 4). Most of these sequences have not been experimentally examined, but virtually all of these organisms have demonstrated acidocalcisomes [4]. Therefore characterization and manipulation of the Phm/Vtc homologues in these various organisms will be an efficient way to explore the biogenesis of acidic organelles in these organisms.
Online data
Acknowledgments
We thank Dr George A. M. Cross (Laboratory of Molecular Parasitology, Rockefeller University, New York, NY, U.S.A.) for providing trypanosome strain 29–13 and plasmid pUB39, Dr John Donelson (Department of Biochemistry, University of Iowa, Iowa City, IN, U.S.A.) for the p2T7Ti vector, Dr Jay Bangs (Department of Biomolecular Chemistry, University of Wisconsin, Madison, WI, U.S.A.) for antibodies against BiP, Dr Norbert Bakalara (Department of Biochimie, Ecole Nationale Supérieure de Chemie de Montpellier, France) for antibodies against TbVP1 and Dr Arthur Kornberg (Department of Biochemistry, Stanford University, Stanford, CA, U.S.A.) for E. coli strain CA38pTrcPPX1. Preliminary sequence data for T. brucei was obtained from the Institute of Genomic Research website. This work was supported by U.S. National Institutes of Health Grant AI-68647 to R. D. K. M. was supported by the Programa de Nanociencia e Nanotecnologia, MCT/CNPq, Brazil and by a training grant from the Ellison Medical Foundation to the Center for Tropical and Emerging Global Diseases.
References
- 1.Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vercesi A. E., Moreno S. N., Docampo R. Ca2+/H+ exchange in acidic vacuoles of Trypanosoma brucei. Biochem. J. 1994;304:227–233. doi: 10.1042/bj3040227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Docampo R., Scott D. A., Vercesi A. E., Moreno S. N. Intracellular Ca2+ storage in acidocalcisomes of Trypanosoma cruzi. Biochem. J. 1995;310:1005–1012. doi: 10.1042/bj3101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Docampo R., de Souza W., Miranda K., Rohloff P., Moreno S. N. Acidocalcisomes: conserved from bacteria to man. Nat. Rev. Microbiol. 2005;3:251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 5.Docampo R., Moreno S. N. Current chemotherapy of human African trypanosomiasis. Parasitol. Res. 2003;90(Suppl. 1):S10–S13. doi: 10.1007/s00436-002-0752-y. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama M., Crooke E., Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 7.Zhang H., Ishige K., Kornberg A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16678–16683. doi: 10.1073/pnas.262655199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Garcia M. R., Kornberg A. Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15876–15880. doi: 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa N., DeRisi J., Brown P. O. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen A., Perzov N., Nelson H., Nelson N. A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 1999;274:26885–26893. doi: 10.1074/jbc.274.38.26885. [DOI] [PubMed] [Google Scholar]
- 11.Nelson N., Perzov N., Cohen A., Hagai K., Padler V., Nelson H. The cellular biology of proton-motive force generation by V-ATPases. J. Exp. Biol. 2000;203:89–95. doi: 10.1242/jeb.203.1.89. [DOI] [PubMed] [Google Scholar]
- 12.Murray J. M., Johnson D. I. Isolation and characterization of Nrf1p a novel negative regulator of the Cdc42p GTPase in Schizosaccharomyces pombe. Genetics. 2000;154:155–165. doi: 10.1093/genetics/154.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray J. M., Johnson D. I. The Cdc42p GTPase and its regulators Nrf1p and Scd1p are involved in endocytic trafficking in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 2001;276:3004–3009. doi: 10.1074/jbc.M007389200. [DOI] [PubMed] [Google Scholar]
- 14.Muller O., Bayer M. J., Peters C., Andersen J. S., Mann M., Mayer A. The Vtc proteins in vacuole fusion: coupling NSF activity to V0 trans-complex formation. EMBO J. 2002;21:259–269. doi: 10.1093/emboj/21.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirtz E., Leal S., Ochatt C., Cross G. A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 16.Lemercier G., Dutoya S., Luo S., Ruiz F. A., Rodrigues C. O., Baltz T., Docampo R., Bakalara N. A vacuolar-type H+-pyrophosphatase governs maintenance of functional acidocalcisomes and growth of the insect and mammalian forms of Trypanosoma brucei. J. Biol. Chem. 2002;277:37369–37376. doi: 10.1074/jbc.M204744200. [DOI] [PubMed] [Google Scholar]
- 17.LaCount D. J., Barrett B., Donelson J. E. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 2002;277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- 18.Rohloff P., Montalvetti A., Docampo R. Acidocalcisomes and the contractile vacuole complex are involved in osmoregulation in Trypanosoma cruzi. J. Biol. Chem. 2004;279:52270–52281. doi: 10.1074/jbc.M410372200. [DOI] [PubMed] [Google Scholar]
- 19.Luo S., Rohloff P., Cox J., Uyemura S. A., Docampo R. Trypanosoma brucei plasma membrane-type Ca2+-ATPase 1 (TbPMC1) and 2 (TbPMC2) genes encode functional Ca2+-ATPases localized to the acidocalcisomes and plasma membrane, and essential for Ca2+ homeostasis and growth. J. Biol. Chem. 2004;279:14427–14439. doi: 10.1074/jbc.M309978200. [DOI] [PubMed] [Google Scholar]
- 20.Montalvetti A., Rohloff P., Docampo R. A functional aquaporin co-localizes with the vacuolar proton pyrophosphatase to acidocalcisomes and the contractile vacuole complex of Trypanosoma cruzi. J. Biol. Chem. 2004;279:38673–38682. doi: 10.1074/jbc.M406304200. [DOI] [PubMed] [Google Scholar]
- 21.Luo S., Fang J., Docampo R. Molecular characterization of Trypanosoma brucei P-type H+-ATPases. J. Biol. Chem. 2006;281:21963–21973. doi: 10.1074/jbc.M601057200. [DOI] [PubMed] [Google Scholar]
- 22.Ault-Riché D., Fraley C. D., Tzeng C. M., Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shatton J. B., Ward C., Williams A., Weinhouse S. A microcolorimetric assay of inorganic pyrophosphatase. Anal. Biochem. 1983;130:114–119. doi: 10.1016/0003-2697(83)90657-7. [DOI] [PubMed] [Google Scholar]
- 24.de Koning H. P., Watson C. J., Jarvis S. M. Characterization of a nucleoside/proton symporter in procyclic Trypanosoma brucei brucei. J. Biol. Chem. 1998;273:9486–9494. doi: 10.1074/jbc.273.16.9486. [DOI] [PubMed] [Google Scholar]
- 25.Rohloff P., Rodrigues C. O., Docampo R. Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol. Biochem. Parasitol. 2003;126:219–230. doi: 10.1016/s0166-6851(02)00277-3. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues C. O., Scott D. A., Docampo R. Characterization of a vacuolar pyrophosphatase in Trypanosoma brucei and its localization to acidocalcisomes. Mol. Cell. Biol. 1999;19:7712–7723. doi: 10.1128/mcb.19.11.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uttenweiler A., Schwarz H., Neumann H., Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell. 2007;18:166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer A., Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J. Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer A., Wickner W., Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 30.Ungermann C., Wickner W., Xu Z. Vacuole acidification is required for trans-SNARE pairing, LMA1 release, and homotypic fusion. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11194–11199. doi: 10.1073/pnas.96.20.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz F. A., Rodrigues C. O., Docampo R. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 2001;276:26114–26121. doi: 10.1074/jbc.M102402200. [DOI] [PubMed] [Google Scholar]
- 32.Scott D. A., Moreno S. N., Docampo R. Ca2+ storage in Trypanosoma brucei: the influence of cytoplasmic pH and importance of vacuolar acidity. Biochem. J. 1995;310:789–794. doi: 10.1042/bj3100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Salcedo J. A., Perez-Morga D., Gijon P., Dilbeck V., Pays E., Nolan D. P. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L., Liang X. H., Uliel S., Unger R., Ullu E., Michaeli S. RNA interference of signal peptide-binding protein SRP54 elicits deleterious effects and protein sorting defects in trypanosomes. J. Biol. Chem. 2002;277:47348–47357. doi: 10.1074/jbc.M207736200. [DOI] [PubMed] [Google Scholar]
- 35.Johnson D. I. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merla A., Johnson D. I. The Cdc42p GTPase is targeted to the site of cell division in the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 2000;79:469–477. doi: 10.1078/0171-9335-00073. [DOI] [PubMed] [Google Scholar]
- 37.Richman T. J., Sawyer M. M., Johnson D. I. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryotic Cell. 2002;1:458–468. doi: 10.1128/EC.1.3.458-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller O., Johnson D. I., Mayer A. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 2001;20:5657–5665. doi: 10.1093/emboj/20.20.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.