Abstract
Defensins are a predominant class of antimicrobial peptides, which act as endogenous antibiotics. Defensins are classified into three distinct sub-families: θ-, β-, and α-defensins. Synthesis of α-defensin has been confirmed only in primates and glires to date and is presumably unique for a few tissues, including neutrophils and Paneth cells of the small intestine. Antimicrobial activities of these peptides were shown against a wide variety of microbes including bacteria, fungi, viruses and protozoan parasites. In the present study, we report the characterization of the equine α-defensin DEFA (defensin α) 1. Transcription analysis revealed that the transcript of the gene is present in the small intestine only. An alignment with known α-defensins from primates and glires displayed a homology with Paneth-cell-specific α-defensins. DEFA1 was recombinantly expressed in Escherichia coli and subsequently analysed structurally by CD and molecular modelling. To examine the antimicrobial properties, a radial diffusion assay was performed with 12 different micro-organisms and the LD90 (lethal dose killing ≥90% of target organism) and MBC (minimal bactericidal concentration) values were examined. DEFA1 showed an antimicrobial activity against different Gram-positive and Gram-negative bacteria and against the yeast Candida albicans. Using viable bacteria in combination with a membrane-impermeable fluorescent dye, as well as depolarization of liposomes as a minimalistic system, it became evident that membrane permeabilization is at least an essential part of the peptide's mode of action.
Keywords: antimicrobial peptide, defensin, horse, innate immunity
Abbreviations: cfu, colony forming unit; DEFA, defensin α; DEFA5L, DEFA 5-like; HD, human defensin; HNP, human neutrophil peptide; LD90, lethal dose killing ≥90% of target organism; MALDI–TOF MS, matrix-assisted laser-desorption ionization–time-of-flight MS; MBC, minimal bactericidal concentration; Ni-NTA, Ni2+–nitrilotriacetate; NP, neutrophil peptide; RACE, rapid amplification of cDNA ends; RP-HPLC, reversed-phase HPLC; TFA, trifluoroacetic acid; TSB, trypticase soy broth
INTRODUCTION
An important component of the innate immunity, next to the complement pathway and phagocytes, are antimicrobial peptides [1,2], which play a pivotal role as key effectors of the immune system and act as endogenous antibiotics [2,3]. Antimicrobial peptides are synthesized by circulating phagocytic cells, leucocytes and epithelial cells of mucosal tissues. In mammals, defensins are a predominant class of these antimicrobial peptides that exhibit a direct activity against a broad range of bacteria [4], fungi [5] and enveloped viruses [6].
Defensins are cationic and cysteine-rich peptides with molecular masses ranging from 3 to 5 kDa. They contain six highly conserved cysteine residues forming characteristic intramolecular disulfide bonds. The disulfide array is specific for the three defensin sub-families in mammals: α-, β-, and θ-defensins [7]. Expression of the α-defensin gene has been confirmed only in primates and glires to date. They were found initially in human neutrophil granulocytes, and identified as natural peptide antibiotics [4]. Further studies showed expression of the gene in mice [8], rhesus macaques [9], rats [10], rabbits [11], guinea pigs [12] and hamsters [13]. Additionally, α-defensin synthesis is presumably unique to a few tissues. The peptides were found in neutrophils, epithelial cells of the human female urogenital tract [14], the kidney of rabbits [15] and in the Paneth cells of different species [16]. Paneth cells are secretory epithelial cells, which are most abundant as specialized ileal cells in the distal small intestine at the base of the crypts of Lieberkuhn. Human Paneth cells synthesize two different α-defensins, HD (human defensin) 5 [DEFA (defensin α) 5] and HD6 (DEFA6) [17,18], in addition to the neutrophil human α-defensin peptides 1–4, also known as HNP [human NP (neutrophil peptide)]-1–4. All α-defensins are synthesized in vivo as inactive precursor proteins. Proteolytic excision of the N-terminal inhibitory anionic propeptide is required for maturation and activation. The signal peptides and propeptides are necessary for correct subcellular sorting and trafficking of the defensins [19,20], and the propeptide is thought to be the major contributing factor that inhibits the antimicrobial activity by neutralization of cationic charges of the mature peptide [20].
The active mature α-defensin peptides consist of 29–35 amino acid residues with a molecular mass of 3–4 kDa. The primary structure shows highly conserved residues, which are indispensable for the structural stability of the peptides. Among them are six invariant cysteine residues, necessary for the typical α-defensin intramolecular disulfide-bond connectivity (Cys1–Cys6, Cys2–Cys4 and Cys3–Cys5); two charged amino acid residues, Arg5, and Glu13, forming a conserved salt bridge [21], and Gly17, which constitutes a signature structural motif which is essential for correct folding [22]. The tertiary structure is a triple-stranded β-sheet with a β-hairpin that contains cationic amino acid residues [23].
The antimicrobial activity of α-defensins was shown against the Gram-negative bacteria Escherichia coli, Salmonella typhimurium and Enterobacter aerogenes; the Gram-positive bacteria Staphylococcus aureus, Bacillus cereus and Listeria monocytogenes; the fungal pathogens Candida albicans and Cryptococcus neoformans; and against the protist Giardia lamblia [9,24–28]. Furthermore, antiviral activity, particularly an anti-HIV-1 activity, was observed [29]. Defensins are thought to kill bacteria by an initial electrostatic interaction with the negatively charged phospholipids of the microbial cytoplasmatic membrane, followed by membrane permeabilization [30,31] and lysis of the microbes [32]. An amphipathic character of the peptide is essential for insertion into the membrane [33].
In the present study, we report the transcription of an equine α-defensin gene in the intestine. The peptide was recombinantly expressed in E. coli and its biological activity against various micro-organisms was determined. In particular, the mechanism of killing was examined by monitoring the membrane-permeabilizing activity of the equine defensin against viable bacteria and liposomes.
EXPERIMENTAL
Transcriptional analysis
On the basis of sequence information for the equine BAC clone CHORI 241-245H5 (GenBank® Nucleotide Sequence Database accession number AY170305 [34]), primers were constructed to amplify a cDNA representing the potential α-defensin. Total RNA was isolated from 13 epithelial tissues (skin, teat, vagina, tongue, gingiva, oesophagus, trachea, lung, small intestine, large intestine, umbilical cord, uterus and meninges) and blood using the RNeasy mini kit (Qiagen). The cDNA was generated by performing RT (reverse transcription)-PCR using an oligo-dT primer OligoT-Bio [5′-ACTCTATGAGAATTCGATGAGCGATCTGT25V]. PCR reactions were performed using the forward primer U130_L1 (5′-ATCTCCTTTGCAGGGGATGAAC) paired with the reverse primer L282_L1 (5′-ACGACAGCAGAGCCTGTAAATGA) and the cDNA as template. The reaction mixtures were incubated at 94 °C for 3 min, followed by 33 cycles at 60 °C for 50 s, 72 °C for 50 s and 94 °C for 1 min. Finally, agarose gel electrophoresis was performed to determine the presence of transcription products.
Molecular cloning and sequencing of DEFA1
The generated product of the small intestine was extracted from the gel using the MinElute gel extraction kit (Qiagen) and purified with the MinElute reaction cleanup kit (Qiagen). The sequences were determined by the DLMBC (Dienstleistungen in der Molekularbiologie und Biochemie) using the forward primer and reverse primer previously used for expression analysis. The resulting sequences revealed sequence overlays (results not shown). New primers were generated: the forward primer DEFA5L (DEFA 5-like)_up1 [5′-TTGACTCCCAGCCATGAG (coding sequence is underlined)] and the reverse primer TailPrimer3′ [5′-ACTCTATGAGAATTCGATGAGCGATCTG (complementary to the OligoT-Bio primer)] to determine the complete cDNA sequence. A 3′-RACE (rapid amplification of cDNA ends) [35] reaction was performed with cDNA from the small intestine as template under the following PCR conditions: 94 °C for 3 min, followed by 33 cycles at 58 °C for 50 s, 65 °C for 45 s and 94 °C for 1 min. The amplified product was purified using the MinElute gel extraction kit (Qiagen). For sequencing, 68 ng of the purified product was subcloned into the pDRIVE cloning vector according to the manufacturer's instructions (Qiagen). A test PCR with the forward primer DEFA5L_up1 and the reverse primer DEFA5L_lo300 (5′-ACGACAGCAGAGCCTGTAAATG) was performed with 20 of the grown clones as a template. Positive clones were sequenced by the DLMBC. The sequences obtained were aligned using the ClustalW program (www.ebi.ac.uk/clustalw/) and analysed.
Preparation of the cDNA for recombinant peptide expression
The cDNA coding for the mature peptide of α-defensin, corresponding to nucleotides 193–296 of the equine α-defensin cDNA (GenBank® Nucleotide Sequence Database accession number EF379126), was amplified using the forward primer DEA6_mat_U2 (5′-GGTATTGAGGGTCGCTCCTGCACCTGCAGACGT, the peptide-specific sequence is underlined) paired with the reverse primer DEA6_mat_L1 (5′-AGAGGAGAGTTAGAGCCTCAGCGACGACAGCAGAG, the peptide-specific sequence is underlined) and the pDRIVE cloning vector containing the complete cDNA as template. The reaction was performed by incubating the reaction mixture at 94 °C for 3 min, followed by 33 cycles at 61 °C for 30 s, 68 °C for 1 min and 94 °C for 45 s. The sample was purified using the MinElute gel extraction kit (Qiagen). The purified PCR products (0.2 pM) were treated for 30 min at 22 °C with 0.16 unit of T4 DNA polymerase (Qiagen) and 3 mM dGTP to generate the overhangs for the annealing reaction. The product was ligated into the vector pET-30 Xa/LIC (Novagen).
Recombinant expression of DEFA1
The vector was transformed into the E. coli strain BL21 (DE3) by heat–shock transformation (90 s at 42 °C). The cells were grown at 37 °C to an attenuance (D600) of 0.5 in 1.5 litres of LB (Luria–Bertani) medium (Roth) containing 30 μg/ml of kanamycin [36]. The expression of the fusion petide was induced with 1 mM IPTG (isopropyl β-D-thiogalactoside), followed by an incubation for 4 h at 37 °C. Bacterial cells were harvested by centrifugation at 6000 g for 20 min at 4 °C and lysed by resuspending the bacterial cell pellet in 40 ml of 6 M guanidine hydrochloride in 100 mM Tris/HCl, pH 8.0, followed by sonication at 70% power for 3 min (Sonopuls; Bandelin). The lysate was clarified by centrifugation at 30000 g for 30 min at 4 °C and the supernatant was sterilized using a membrane filter with a 0.45 μm pore size (Sartorius) prior to peptide purification.
Purification of the recombinant peptide
The His6-tagged α-defensin fusion peptide was purified by performing Ni–NTA (Ni2+–nitrilotriacetate) metal-affinity chromatography using prepacked Ni–NTA superflow columns (Qiagen). Clarified lysates were transferred to the column and drained by gravity flow. Washing was performed with 6 M guanidine hydrochloride in 100 mM Tris/HCl, pH 8.0, and 15 mM imidazole, and the His6-tagged peptide was eluted with 6 M guanidine hydrochloride in 100 mM Tris/HCl, pH 8.0, and 750 mM imidazole. The samples were stored at −20 °C.
The buffer of the dissolved fusion peptides was exchanged by using PD-10 desalting columns (GE Healthcare) with a bed volume of 8.3 ml. Ni–NTA-purified peptides were transferred to a PD-10 column and eluted using cleavage buffer consisting of 20 mM Tris/HCl, pH 6.5, 50 mM NaCl and 1 mM CaCl2. The buffer-exchange procedure was repeated twice. The eluted His6-tagged peptide was quantified photometrically by measuring the UV absorption at 280 nm on the basis of the molar absorption coefficient [37].
The fusion peptide was cleaved for 12 h at room temperature (20 °C) with 10 units of Factor Xa protease (Qiagen) per 150 μg of peptide in cleavage buffer.
Reversed-phase chromatography
After cleavage, the mature peptide was purified immediately to apparent homogeneity by using RP-HPLC (reversed-phase HPLC). The sample was applied to a C18 preparative RP-HPLC VP 250/10 Nucleosil 300-7 column (Macherey–Nagel) and the peptide was eluted after 16 min using a 22–40% (v/v) acetonitrile gradient developed over 90 min. Finally, the peptide was freeze-dried and redissolved in 0.01% (v/v) TFA (trifluoroacetic acid) for storage at −20 °C and further studies.
CD spectroscopy
CD measurements were carried out on a JASCO J-720 spectropolarimeter (Japan Spectroscopic Company) calibrated as described by Chen and Yang [38]. The spectral bandwidth was 2 nm. Three scans were recorded between 190 nm and 250 nm, with 1 nm acquisition steps, and the spectrum was baseline corrected. Measurements were carried out at room temperature and cuvettes of 1.0 mm optical pathlength were used. The peptide concentration was 15 μM in 0.01% (v/v) TFA.
MS analysis
The molecular identity and homogeneity of the preparation was verified by MALDI–TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS). The samples were mixed with a saturated matrix solution of sinapinic acid prepared in 80% (v/v) acetonitrile/0.1% (v/v) TFA and spotted on a stainless steel sample target. Mass spectra were obtained on a MALDI–TOF/TOF mass spectrometer (4700 Proteomics Analyser; Applied Biosystems) in the positive-ion linear mode and calibrated externally. The average mass of the proteins were compared with the theoretical average mass of DEFA1 considering potential disulfide formation.
Antimicrobial activity
The following bacterial strains were used in a radial diffusion assay as described by Lehrer et al. [39]: Gram-negative E. coli A.T.C.C. 11303, A.T.C.C. 11775, A.T.C.C. 25922, A.T.C.C. 35218 and D31; Pseudomonas aeruginosa A.T.C.C. 10145; Burkholderia cepacia A.T.C.C. 25416 and Gram-positive Staph. aureus A.T.C.C. 6538 and A.T.C.C. 12600; Staph. epidermidis A.T.C.C. 14990; Bacillus megaterium A.T.C.C. 14581 and the yeast C. albicans A.T.C.C. 24433. The organisms were grown to mid-exponential phase in TSB (trypticase soy broth), suspended into agar [1% (w/v) agarose in 10 mM sodium phosphate, pH 7.4] supplemented with a 1:100 dilution of 1×TSB, and poured into Petri dishes (10 ml). After hardening, 5 μl of the peptide test solution was pipetted into wells formed using a biopsy punch (3 mm diameter) and allowed to incubate at 37 °C overnight. Lysozyme was used as a control peptide and 0.01% (v/v) TFA served as a negative control. Plates were then overlayed with 10 ml of 1% (w/v) agar supplemented with double-strength TSB [3.4% (w/v) casein peptone, 0.6% (w/v) tryptic soy peptone, 0.5% (w/v) glucose, 1% (w/v) NaCl and 0.5% (w/v) K2HPO4]. After incubation for 3 h, zones of growth inhibition surrounding the wells were measured.
Additionally, LD90 (lethal dose killing ≥90% of target organism) and MBC (minimal bactericidal concentration, ≥99.9% killing of the target organism) values for the peptide were determined by using the test-strain pattern described for the radial diffusion assay, with the exception of E. coli A.T.C.C. 11030. In addition, Gram-negative Klebsiella pneumoniae A.T.C.C. 13883; Enterobacter cloacae A.T.C.C. 13047; P. aeruginosa A.T.C.C. 11440 and Gram-positive Enterococcus faecalis A.T.C.C. 29212 and PEG (Paul-Ehrlich-Gesellschaft) 205 (wild-type) and Streptococcus pyogenes A.T.C.C. 12344 were introduced. Briefly, after a 2–3 h growth period in brain–heart infusion broth at 36±1 °C, the bacteria were washed three times in 10 mM sodium phosphate buffer, pH 7.4, and adjusted to 104–105 cfu (colony forming units)/ml in the same buffer. The peptide solution (10 μl; range of final concentrations tested 0.05–6.1 μM) was added to 100 μl of the bacterial suspension and incubated at 36±1 °C for 2 h before colony formation was determined. The peptide solvent [0.01% (v/v) TFA] served as a negative control.
Permeabilization of bacterial membranes
Bacteria with compromized membranes were detected by monitoring the fluorescence of the DNA-binding dye SYTOX Green (Molecular Probes). Bac. megaterium A.T.C.C. 14581 in midexponential phase was washed twice and resuspended in 10 mM Hepes and 25 mM NaCl, pH 7.4. A flat-bottom 96-well microtitre plate (Sarstedt) was coated with 0.1% (w/v) BSA (A2153, Sigma–Aldrich) for 15 min prior to use in the assay. Peptides were 2-fold serially diluted in 10 mM Hepes and 25 mM NaCl, pH 7.4. Bacteria (1×105 cfu/50 μl) were incubated with the diluted peptides (25 μl) and 2 μM of the fluorescent dye SYTOX Green (25 μl; in 10 mM Hepes and 25 mM NaCl, pH 7.4) at 20 °C for 1 h. Permeabilization of the bacterial cytoplasmic membrane allowed the dye to cross this membrane and to intercalate with the DNA. When excited at 495 nm, the binding of the dye to DNA resulted in an increase of emitted fluorescence at 538 nm, which was measured in a microtitre plate reader (Fluoroskan II; Labsystems). Membrane-permeabilising activity of the peptides was expressed as a percentage of permeabilized bacteria. To monitor the activity at pH 5.2, 20 mM Mes and 25 mM NaCl was used. As a control antimicrobial peptide, cecropin P1 (HPLC-grade; Sigma–Aldrich) was used in parallel. For maximum permeabilization of the bacteria (100%), cells were incubated with 70% (v/v) ethanol for 10 min. The values are expressed as means (n=2), with each experiment performed in duplicate.
Assay for pore-forming activity
Pore-forming activity of samples was determined by measuring fluorimetrically the dissipation of a valinomycin-induced membrane potential in liposomes prepared from crude soy bean phospholipids [40]. Fluorescence was measured by a fluorescence spectrophotometer (model LS 50B; PerkinElmer) using excitation and emission wavelengths of 620 nm and 670 nm respectively. Pore-forming activity was measured as the initial change in fluorescence intensity over time after addition of the sample. As a control pore-forming peptide, alamethicin (in HPLC-grade; Sigma–Aldrich) was used in parallel.
Molecular modelling
The three-dimensional model for equine α-defensin was generated using the X-ray structure of the human α-defensin HNP-3 (PDB ID: 1DFN) as a template. According to the alignment, amino acid residues were exchanged in the template structure. Insertions and deletions were modelled by using a database search approach included in the software package WHAT IF [41]. The database was searched for a peptide sequence of the appropriate length, which was fitted to the template. All loops were selected from the database so as to give a minimum root mean square distance between the ends of the loops and helices. Loops with unfavourable backbone dihedral angles or van der Waals clashes were excluded. Finally, these model structures were energy minimized, using the steepest descent algorithm implemented in the GROMOS force field [42]. The structural representation was performed using the RIBBON program [43].
RESULTS
Transcriptional analysis
A single amplified product could be verified in the small intestine of the horse (Figure 1). This analysis failed to detect gene transcription in skin, teat, vagina, tongue, gingiva, oesophagus, trachea, lung, large intestine, umbilical cord, uterus, meninges and blood. The amplified product of the horse small intestine was approx. 340 bp long. The specific transcription of the equine α-defensin-like gene suggested the existence of an α-defensin in the intestine of the horse.
Figure 1. Transcriptional analysis of DEFA1 in various equine tissues.
DEFA1 is solely transcribed in the small intestine (lane 9, lower panel). The product size is approx. 340 bp in length. β-Actin was used as a positive control (upper panel), water was used instead of cDNA template as a negative control (lane 15).
Determination of the primary structure of the DEFA1 precursor
Sequence overlays were determined by sequencing of the amplified product from the gut with primers used for transcriptional analysis. It could not be excluded that more than one product has been amplified. Therefore subcloning was performed with amplified products generated by 3′-RACE to determine the complete coding sequence of the amplified product. According to the transcriptional analysis, cDNA of the small intestine was used for the PCR reaction. Positive clones were sequenced with forward and reverse primers used for 3′-RACE. Two distinct cDNA sequences, DEFA5L (already known from the BAC clone CHORI 241-245H5), and a new sequence, DEFA1 (GenBank® Nucleotide Sequence Database accession number EF379126) were identified.
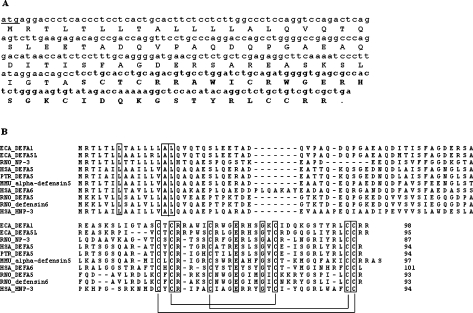
The cDNA sequence of the DEFA1 gene possessed an open reading frame of 297 nucleotides including the start and stop codons (Figure 2A). In comparison with sequences available in public databases, the region of the putative propetide and mature peptide was predicted. The putative coding region of both the signal peptide and the propeptide is 192 nucleotides in length, the coding region of the mature peptide consists of 105 nucleotides. ClustalW alignment of the DEFA1 amino acid sequence with the equine DEFA5L sequence (NCBI accession number CAJ01789) and the sequences of human DEFA5 (NCBI accession number EAW80489), DEFA6 (NCBI accession number AAC50382) and HNP-3 (NCBI accession number NP_005208), chimpanzee DEFA5 (NCBI accession number NP_001012657), rhesus macaque α-defensin 5 (NCBI accession number AAW51369); rat DEFA5 (NCBI accession number P82106), α-defensin 6 (NCBI accession number AAT91348) and NP-3 (NCBI accession number Q62713) showed a relatively conserved region in the signal peptide and a more divergent region in the mature peptide (Figure 2B). Equine DEFA5L had the highest identity with DEFA1, with 84% amino-acid-sequence identity. DEFA1 had 48% amino acid sequence identity with human and chimpanzee DEFA5 and with rat NP-3. The rhesus macaque α-defensin 5 showed an amino acid sequence identity of 44% with DEFA1; and the rat Paneth-cell-specific α-defensins DEFA5 and defensin 6 had an amino acid identity of 39% with DEFA1. Human DEFA6 had 37% amino acid identity with DEFA1, whereas the protein with the lowest identity with DEFA1 was human HNP-3 (35%). The six cysteine residues (Cys2, Cys4, Cys10, Cys20, Cys31 and Cys32) are completely conserved within the peptides. Equally, the arginine and glutamine residues (Arg5 and Glu13), which are necessary for the intramolecular salt bridge, are conserved, as well as the glycine residue at position 17 (Gly17), which is essential for correct folding. The putative mature peptide (34 amino acid residues in length) starts with a serine residue, followed by the first cysteine residue. The complete primary structure of the DEFA1 precursor consists of 98 amino acid residues, including the signal peptide and propeptide.
Figure 2. DEFA1 nucleotide sequence and the amino acid sequence and alignment of the DEFA1 primary structure with those of other α-defensins.
(A) The DEFA1 cDNA sequence is depicted in lower case letters, with the amino acid sequence in upper case letters. The sequence (nucleotides and amino acids) of the mature peptide is depicted in bold, and the start codon is underlined. (B) Comparison of the DEFA1 amino acid sequence with the amino acid sequences of equine [ECA (Equus caballus)] DEFA5L, human [HSA (Homo sapiens)] α-defensins DEFA5, DEFA6 and HNP-3, chimpanzee [PTR (Pan troglodytes)] DEFA5, rat [RNO (Rattus norwegicus)] DEFA5, NP-3 and defensin 6, and rhesus macaque [MMU (Macaca mulatta)] α-defensin 5. The boxes indicate highly conserved residues among the primary translation products of equine defensin and the other α-defensin genes. The connectivity of the cysteines typically present in α-defensins is indicated by lines. The length of the peptides is denoted at the C-terminus.
Recombinant expression and purification of DEFA1
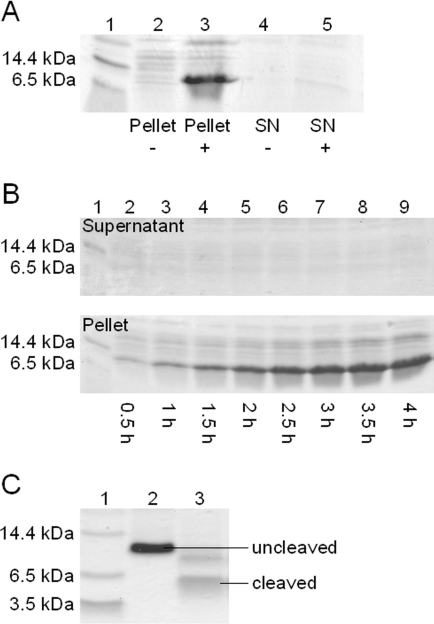
Expression of recombinant DEFA1 was performed by cloning the cDNA sequence of the mature α-defensin peptide (Figure 2A) into the pET30-Xa/LIC vector and transfecting E. coli BL21 (DE3) cells with the vector. The fusion peptide was exclusively synthesized in inclusion bodies and therefore was only detected in the pellet of cells after cell lysis and centrifugation at 30000 g (Figure 3A). Analysis of bacterial extracts by SDS/PAGE at different time points revealed peptide synthesis in inclusion bodies already occurring during the first 30 min after induction of gene expression. A further increase in peptide concentration after induction was observed after an expression time of up to 4 h (Figure 3B). The recombinant product was confirmed by SDS/PAGE to have a molecular mass of approx. 10 kDa, close to the theoretical mass of 9.046 kDa. After Ni–NTA acid metal-affinity chromatography, the cleavage of the product with Factor Xa resulted in a peptide of approx. 5 kDa as determined by Tricine/PAGE (Figure 3C). This result is in good agreement with the expected molecular mass of 4.071 kDa.
Figure 3. Recombinant expression of DEFA1.
(A) SDS/PAGE of the cell pellet before induction (lane 2) and after induction (lane 3) and the supernatant from unstimulated bacterial cells (lane 4) and stimulated bacterial cells (lane 5). Lane 1, molecular-mass markers. The His6-tagged fusion peptide was only detected in the pellet of stimulated cells, and has a molecular mass of approx. 10 kDa. SN, supernatant. (B) Time-dependent analysis of peptide expression after induction in the supernatant (upper panel) and the pellet (lower panel) fractions after 0.5 h (lane 2), 1 h (lane 3), 1.5 h (lane 4), 2 h (lane 5), 2.5 h (lane 6), 3 h (lane 7), 3.5 h (lane 8) and 4 h (lane 9). The molecular-mass markers are shown in lane 1. (C) Digestion of the recombinant peptide with Factor Xa. Lane 1, molecular-mass markers; lane 2, the uncleaved peptide with a mass of approx. 10 kDa; lane 3, cleaved product. The mature peptide reveals a molecular mass of approx. 5 kDa. SN, supernatant.
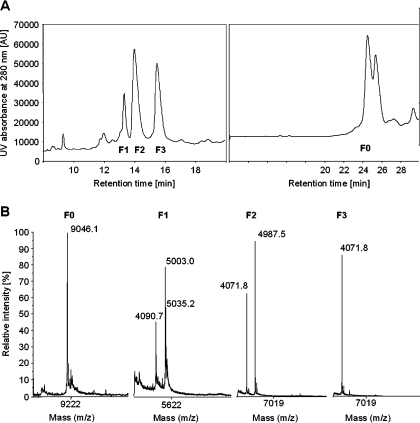
Recombinant DEFA1 was purified to homogeneity by performing RP-HPLC. Three conspicuous peaks appeared at elution times of 13.4, 14.0 and 15.7 min (Figure 4A). The fractions were analysed by MALDI–TOF MS. The fraction F1 contained an unknown peptide mixture with peptide masses of approx. 4090, 5003 and 5035 Da. Fractions F2 and F3 contained peptides with the mass of 4071.8 Da (Figure 4B). Notably, for the mature DEFA1 peptide, an average mass of 4071.8 Da was calculated, provided that the six cysteine residues were involved in disufide bonds. In fraction F2, the His6-tag (4.987 kDa) was eluted in addition to the mature peptide, whereas in fraction F3, homogeneous material representing the DEFA1 peptide was found. The accurate monoisotopic mass of the peptide was also determined (4068.9 Da) and the molecular identity of DEFA1 was confirmed. F3 eluates were used for further studies. The eluates were freeze-dried and the purified peptide was resuspended in 0.01% (v/v) TFA for further investigation. Finally, MALDI–TOF MS was performed to prove the purity of the DEFA1 peptide (results not shown).
Figure 4. RP-HPLC purification of DEFA1 and MALDI–TOF MS of the eluted fractions.
(A) The chromatogram on the left-hand side shows three conspicuous peaks appearing at elution times of 13.4, 14.0 and 15.7 min. Eluted fractions were termed F1 to F3 and analysed by MS. An RP-HPLC analysis of the undigested peptide is shown on the right-hand side, the eluted fraction was termed F0. (B) MALDI–TOF MS of the fractions F0–F3. Fraction F0 shows a mass peak of 9046.1 Da for the uncleaved product. Fraction F1 shows an unknown peptide mixture with peptide masses of approx. 4090, 5003 and 5035 Da. Fractions F2 and F3 contain peptides of the calculated average mass of cleaved DEFA1 (4071.8 Da), whereas in fraction F2, the His6-tag (4987.5 Da) was eluted in addition to the mature peptide. In fraction F3, a homogenous peptide without any contaminants was detected.
Structural characterization
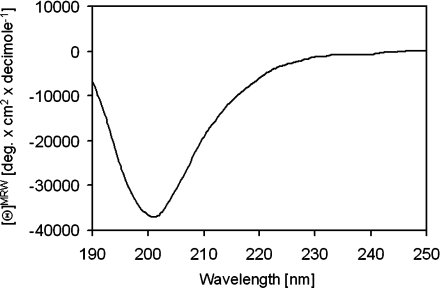
The recombinantly expressed DEFA1 was structurally characterized by CD spectroscopy. The CD spectrum (in the range of 190 nm and 250 nm) is shown in Figure 5 and displays a minimum at 203 nm. The shape of the spectrum is indicative of a β-sheet structure with a high content of P2 elements [44]. This is in agreement with the known α-defensin structures.
Figure 5. CD spectrum of DEFA1.
The spectrum was recorded between 190 nm and 250 nm in 0.01% (v/v) TFA, with a minimum at 203 nm.
Antimicrobial activity of DEFA1
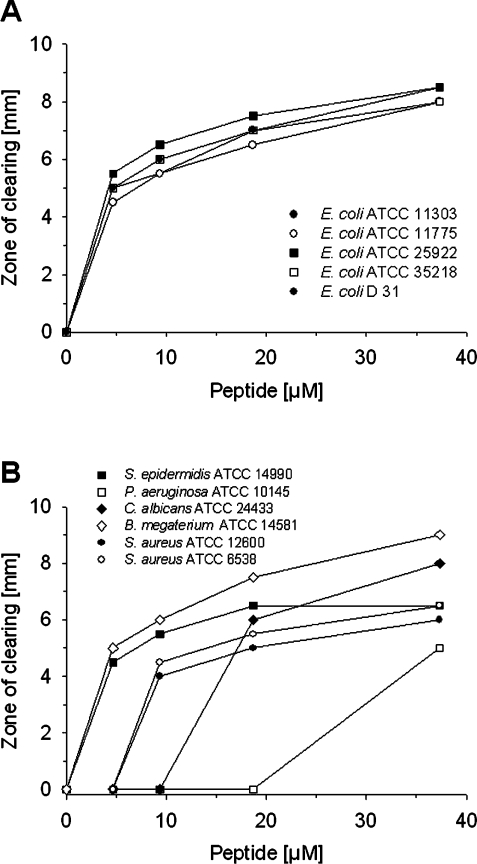
To assess the antimicrobial activity of DEFA1, a radial diffusion assay was performed. As shown in Figure 6, treatment of micro-organisms with the recombinant equine α-defensin resulted in a visible inhibition of growth (zones of clearing) against all of the target organisms with the exception of B. cepacia (results not shown). The relative potential of the peptide varied depending on the micro-organisms tested. Whereas DEFA1 was only lightly active against P. aeruginosa A.T.C.C. 10145, potent activity was found against Bac. megaterium, all E. coli strains, Staph. epidermidis and Staph. aureus strains. Against seven test strains (all E. coli strains, Staph. epidermidis and C. albicans), peptide concentrations as low as 4.7 μM led to growth inhibition zones. The Staph. aureus strains were inhibited at peptide concentrations of ≥9.3 μM and C. albicans was susceptible at ≥18.7 μM.
Figure 6. Antimicrobial activities of DEFA1 in the radial diffusion assay.
Peptide concentration (x-axis) plotted against the diameter of the microbial growth inhibition zone (y-axis) after incubation for 12 h. (A) Growth inhibition of various E. coli strains. (B) Growth inhibition of Staph. aureus, Staph. epidermidis, P. aeruginosa, Bac. megaterium, and C. albicans. The arrangement of measuring points are a result of adjusted peptide concentrations of 4.7, 9.3, 18.7, and 37.3 μM.
Additionally, the minimal concentration of the equine DEFA1 that killed 90% (LD90) and 99.9% (MBC) of 17 target micro-organisms is shown in Table 1. All organisms were tested with various defensin concentrations (0.05–6.1 μM). This range was sufficient to determine the lethal dose of the target organisms. DEFA1 displayed a broad spectrum of antimicrobial activity against both fermenting and non-fermenting Gram-negative strains, against Gram-positive cocci (genera Staphylococcus, Streptococcus and Enterococcus), spore-forming Bac. megaterium and the yeast C. albicans. Bac. megaterium was more susceptible to DEFA1 than any of the other strains tested. In general, E. coli strains were more susceptible to the peptide than other fermenting Gram-negative bacteria such as K. pneumoniae and Eb. cloacae, which were weakly susceptible to the peptide. Non-fermenting Gram-negative bacteria, such as P. aeruginosa and B. cepacia, were weakly susceptible or showed no sensitivity respectively to DEFA1. The Gram-positive cocci, such as Staph. aureus, Staph. epidermidis, Ec. faecalis, and Strep. pyogenes, exhibited a high sensitivity to DEFA1 at reliable peptide concentrations. The yeast C. albicans demonstrated an MBC value of 0.8 μM, whereas B. cepacia was not killed with peptide concentrations of up to 6.1 μM. These results correspond with the results from the radial diffusion assay.
Table 1. LD90 and MBC values for DEFA1 against different micro-organisms.
LD90 and MBC values are defined as the lethal dose of DEFA1 that killed 90% (LD90) or 99.9% (MBC) of the target organisms. The highest peptide concentration tested was 6.1 μM.*, no killing detected at the indicated concentration.
| Test organism | Gram stain reaction | LD90 (μM) | MBC (μM) |
|---|---|---|---|
| E. coli A.T.C.C. 11775 | − | 0.2 | 0.4 |
| E. coli A.T.C.C. 25922 | − | 0.2 | 0.4 |
| E. coli A.T.C.C. 35218 | − | 0.05 | 0.4 |
| E. coli D31 | − | 0.4 | 0.8 |
| K. pneumoniae A.T.C.C. 13883 | − | 3 | >6.1* |
| Eb. cloacae A.T.C.C. 13047 | − | 6.1 | >6.1* |
| P. aeruginosa AT.C.C. 10145 | − | 3 | >6.1* |
| P. aeruginosa A.T.C.C. 11440 | − | 1.5 | 3 |
| B. cepacia A.T.C.C. 25416 | − | >6.1* | >6.1* |
| Staph. aureus A.T.C.C. 6538 | + | 0.4 | 0.8 |
| Staph. aureus A.T.C.C. 12600 | + | 0.4 | 1.5 |
| Staph. epidermidis A.T.C.C. 14990 | + | 0.8 | 1.5 |
| Ec. faecalis A.T.C.C. 29212 | + | 0.2 | 6.1 |
| Ec. faecalis PEG 205 (wild-type) | + | 0.4 | 3 |
| Strep. pyogenes A.T.C.C. 12344 | + | 0.8 | 0.8 |
| Bac. megaterium A.T.C.C. 14581 | + | 0.05 | 0.2 |
| C. albicans A.T.C.C. 24433 | 0.8 | 0.8 |
Characterization of the mode of action of the peptide
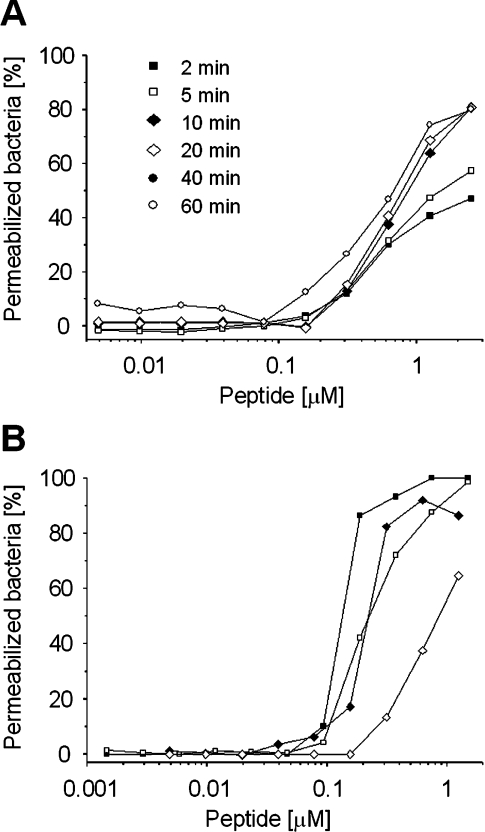
To investigate the mode of action of DEFA1, we used Bac. megaterium and the fluorescent dye SYTOX Green. As soon as the dye enters the bacterial cell it intercalates with the DNA and its fluorescence is enhanced. This assay confirmed that DEFA1 is potently active against these Gram-positives bacteria (Figure 7A). Notably, it demonstrated that the peptide rapidly permeates bacterial membranes. As DEFA1 contains a histidine residue in its primary structure, we evaluated the effect of a more acidic pH at which the histidine residue is protonated on the membrane-permeabilizing activity. The efficacy of horse defensin to induce membrane lesions in viable bacteria was found to be very similar at both pH 5.2 and at pH 7.4, and comparable to that of the well-known antimicrobial peptide cecropin P1 (Figure 7B).
Figure 7. Membrane permeabilization of Bac. megaterium induced by DEFA1.
Membrane damage of the bacteria was measured fluorometrically using the SYTOX Green dye. The binding of the dye to the DNA in membrane-compromised target cells resulted in an increase in fluorescence. Antibacterial activity of the peptides is expressed as a percentage of permeabilized bacteria. (A) Time kinetics of membrane permeabilization induced by DEFA1 measured for different concentrations of peptide at various incubation periods. (B) Comparison of the membrane-permeabilizing effects of DEFA1 (◆,◇) and cecropin P1 (■,□) after incubation of Bac. megaterium for 30 min with each peptide at various concentrations and either at pH 5.2 (◆,■) or at pH 7.4 (◇,□).
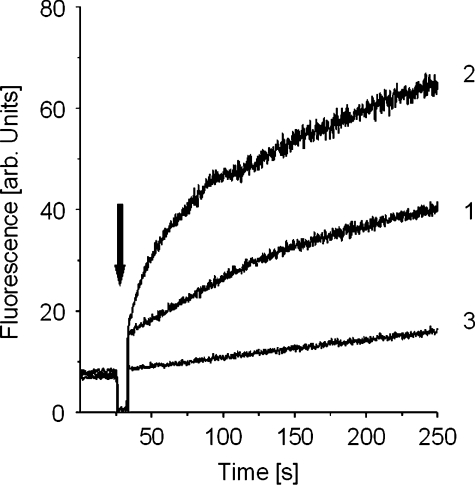
We also measured the pore-forming activity of DEFA1 to further characterize its mode of action by using a minimalistic membrane system. More precisely, we monitored the dissipation of a membrane potential induced in liposomes composed of azolectin, a crude phospholipid mixture from soy bean. It became evident that the equine defensin displayed pore-forming activity (Figure 8), although only moderately when compared with alamethicin, the prototype of a pore-forming peptide which acts according to the barrel–stave model [45].
Figure 8. Time course of pore formation induced by DEFA1.
The dissipation of a valinomycin-induced diffusion potential in vesicles of soy bean phospholipids after addition of DEFA1 (1.4 nmol; arrow) (1), the control peptide alamethicin (0.1 nmol) (2) and the peptide solvent (3) was recorded. Pore-forming activity was reflected by the increase in fluorescence as a function of time.
DISCUSSION
The existence of an α-defensin peptide in the small intestine of the horse was indicated by transcriptional analysis, and the amino acid sequence obtained was aligned with other Paneth-cell-specific α-defensin sequences. Interestingly, except in primates and rodents, expression of α-defensin has not been verified in any other species to date. The horse has now been added to the group of animals expressing α-defensins, and this may lead to a new perception in the evolution of defensins. A possible ancestral gene for DEFA5 may already have been present before euarchontoglires (including glires and primates) and laurasiatheria (including perissodactyla amongst others) evolved to two different subgroups. It is unlikely that the equine DEFA1 identified had developed independently, as indicated by the high identity of the amino acid sequences between equine DEFA1 and other known α-defensins (Figure 2B). This is supported by the finding of genomic α-defensin-like sequences in basal mammals [46].
DEFA1 also retained the typical conservation of a signal-peptide, a negatively-charged prosegment and the α-defensin characteristically-conserved amino acid residues, including the six cysteine residues, Arg5, Glu13 and Gly17. Most α-defensins contain none or only one charged or polar uncharged amino acid residue following the last two cysteines. C-terminal tails which are rich in charged or polar uncharged amino acid residues, are predicted to be exposed to the surface to influence the overall charge and amphipathicity of the peptide. The equine defensins DEFA1 and DEFA5L have two C-terminal arginine residues (Figure 2B). An extended C-terminus is also shown on rhesus macaque α-defensin 5. In previous studies, a general relationship between higher defensin cationicity and a greater microbicidal potency was noted [5]. It was supposed that the net charge covers a different antimicrobial spectrum and efficiency of these peptides, and is strongly correlated with antimicrobial activity of the defensins [47]. Equine DEFA1 is less cationic (+2) than human DEFA5 (+5) and Paneth cell α-defensins from rhesus macaque small intestine (+4.5 to +7.5), but equine DEFA1 showed at least similar antimicrobial activities, suggesting that the relative cationicity did not appear to be a major factor in the killing of bacteria. This result is consistent with those of previous studies demonstrating that the overall microbicidal activity of α-defensins is not merely a function of their net charge [48], but the defensins may possess specifities for bacterial strains.
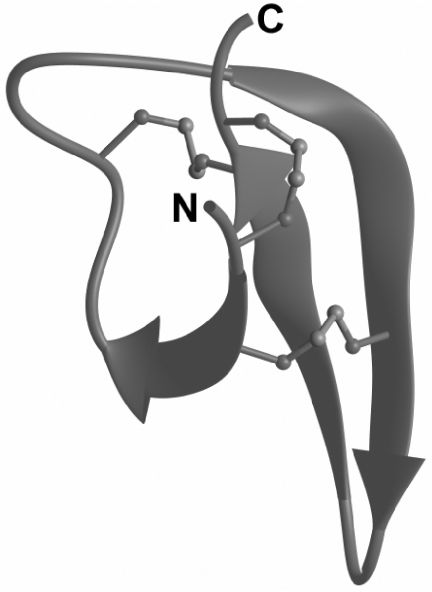
A three-dimensional model of the DEFA1 structure is shown in Figure 9. The structuer is a small β-sheet, consisting of three β-strands flanked by large unstructured regions. The long loop connecting the first and the second β-strand contains several cationic amino acid residues. Together with the two arginines located at the C-terminus, this loop forms an epitope with a high positive-charge density.
Figure 9. Three-dimensional model of DEFA1.
β-Sheet structures are displayed as arrows, the N-terminus is marked with N, the C-terminus with C. Disulfide bonds are depicted as ball-and-stick representations.
Alignment of DEFA1 and DEFA5L with other amino acid sequences revealed homology with human DEFA5. The antimicrobial properties of human DEFA5 have been investigated comprehensively [24]. In summary, it has been established that human DEFA5 efficiently kills Gram-negative and Gram-positive bacteria at comparable peptide concentrations and has shown a consistently high activity against a broad spectrum of micro-organisms, including fungi. Human DEFA5 was potent in almost the same manner as the neutrophil peptides HNP 1–4 against Gram-positive Staph. aureus, but displayed a conspicuously higher activity against E. coli. By comparing the activities of equine DEFA1 against E. coli, Staph. aureus and Staph. epidermidis, we also determined a higher activity against E. coli. Other strains of Gram-negative bacteria such as Eb. cloacae, K. pneumoniae, P. aeruginosa and B. cepacia were weakly sensitive. Similarly, Gram-positive cocci such as Staph. aureus, Staph. epidermidis, Strep. pyogenes and Ec. faecalis are highly sensitive to DEFA1 at peptide concentrations <1 µM (LD90). Collectively, despite the broad antimicrobial spectrum of DEFA1, the most sensitive targets of this peptide are Gram-positive bacteria. It appears that a preference for particular bacteria applies to both human DEFA5 and equine DEFA1.
The mode of action of Paneth cell α-defensins has not been studied to date. For antimicrobial peptides in general, a two-step mechanism has been proposed to explain their killing effect on micro-organisms. Most cationic peptides bind to the target-cell-surface, as a result of the presence of negatively charged phospholipids or other moieties. Many of them subsequently disturb the membrane integrity [2]. Likewise, killing of bacteria by the equine defensin was accompanied by permeabilization of the bacterial cytoplasmic membranes, similar to the action of other antimicrobial peptides, such as cecropin P1. Moreover, in a minimalistic system using the depolarization of liposomes as a measure of pore-forming activity, it became evident that DEFA1 is moderately active in creating pores in artificial membranes, a property described previously for human α-defensins [49].
In recent studies, a specific deficiency of human DEFA5 was observed in patients with ileal Crohn's disease, a chronic disease of the intestine characterized by inflammation of the gut. The horse also displays similar disease patterns, for example in anterior enteritis (duodenitis) or colitis X, which are catarrhal–haemorrhagic inflammations of the gut. It will be interesting to examine whether a possible relationship exists between these infectious diseases and a possible deficiency in equine Paneth cell α-defensins.
Acknowledgments
We thank the BMBF (Bundesministerium für Bildung und Forschung) [FUGATO (Funktionelle GenomAnalyse im Tierischen Organismus)] and the German Research Council [DFG (Deutsche Forschungsgemeinschaft), SFB 617] for supporting this study. We thank Ms Sonja Hollmer from the Institute of Biochemistry, University of Kiel and Ms Heidrun Ließegang from the Zoological Institute, University of Kiel, for technical assistance, and Dr Ulrike Diesterbeck (Institute of Virology, University of Göttingen) for valuable discussions.
References
- 1.Boman H. G. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;40:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 4.Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selsted M. E., Szklarek D., Ganz T., Lehrer R. I. Activity of rabbit leukocyte peptides against Candida albicans. Infect. Immun. 1985;49:202–206. doi: 10.1128/iai.49.1.202-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daher K., Selsted M. E., Lehrer R. I. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 1986;60:1068–1074. doi: 10.1128/jvi.60.3.1068-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang D., Biragyn A., Kwak L. W., Oppenheim J. J. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhauer P. B., Harwig S. S., Lehrer R. I. Cryptdins: antimicrobial defensins of the murine small intestine. Infect. Immun. 1992;60:3556–3565. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y., Yuan J., Miller C. J., Selsted M. E. Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of α-defensins from rhesus macaque leukocytes. Infect. Immun. 1999;67:6139–6144. doi: 10.1128/iai.67.11.6139-6144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yount N. Y., Wang M. S., Yuan J., Banaiee N., Ouellette A. J., Selsted M. E. Rat neutrophil defensins. Precursor structures and expression during neutrophilic myelopoiesis. J. Immunol. 1995;155:4476–4484. [PubMed] [Google Scholar]
- 11.Sinha S., Cheshenko N., Lehrer R. I., Herold B. C. NP-1, a rabbit α-defensin, prevents the entry and intercellular spread of herpes simplex virus type 2. Antimicrob. Agents Chemother. 2003;47:494–500. doi: 10.1128/AAC.47.2.494-500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selsted M. E., Harwig S. S. Purification, primary structure, and antimicrobial activities of a guinea pig neutrophil defensin. Infect. Immun. 1987;55:2281–2286. doi: 10.1128/iai.55.9.2281-2286.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mak P., Wojcik K., Thogersen I. B., Dubin A. Isolation, antimicrobial activities, and primary structures of hamster neutrophil defensins. Infect. Immun. 1996;64:4444–4449. doi: 10.1128/iai.64.11.4444-4449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quayle A. J., Porter E. M., Nussbaum A. A., Wang Y. M., Brabec C., Yip K. P., Mok S. C. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am. J. Pathol. 1998;152:1247–1258. [PMC free article] [PubMed] [Google Scholar]
- 15.Wu E. R., Daniel R., Bateman A. RK-2: a novel rabbit kidney defensin and its implications for renal host defense. Peptides. 1998;19:793–799. doi: 10.1016/s0196-9781(98)00016-3. [DOI] [PubMed] [Google Scholar]
- 16.Ayabe T., Satchell D. P., Wilson C. L., Parks W. C., Selsted M. E., Ouellette A. J. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 17.Jones D. E., Bevins C. L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 18.Jones D. E., Bevins C. L. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 19.Liu L., Ganz T. The pro region of human neutrophil defensin contains a motif that is essential for normal subcellular sorting. Blood. 1995;85:1095–1103. [PubMed] [Google Scholar]
- 20.Valore E. V., Martin E., Harwig S. S., Ganz T. Intramolecular inhibition of human defensin HNP-1 by its propiece. J. Clin. Invest. 1996;97:1624–1629. doi: 10.1172/JCI118588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z., Li X., DeLeeuw E., Ericksen B., Lu W. Why is the Arg5-Glu13 salt bridge conserved in mammalian α-defensins? J. Biol. Chem. 2005;280:43039–43047. doi: 10.1074/jbc.M510562200. [DOI] [PubMed] [Google Scholar]
- 22.Xie C., Prahl A., Ericksen B., Wu Z., Zeng P., Li X., Lu W. Y., Lubkowski J., Lu W. Reconstruction of the conserved β-bulge in mammalian defensins using D-amino acids. J. Biol. Chem. 2005;280:32921–32929. doi: 10.1074/jbc.M503084200. [DOI] [PubMed] [Google Scholar]
- 23.Bals R. Antimicrobial peptides and peptide antibiotics. Med. Klin. 2000;95:496–502. doi: 10.1007/pl00002139. [DOI] [PubMed] [Google Scholar]
- 24.Ericksen B., Wu Z., Lu W., Lehrer R. Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 2004;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouellette A. J., Satchell D. P., Hsieh M. M., Hagen S. J., Selsted M. E. Characterization of luminal paneth cell α-defensins in mouse small intestine. Attenuated antimicrobial activities of peptides with truncated amino termini. J. Biol.Chem. 2000;275:33969–33973. doi: 10.1074/jbc.M004062200. [DOI] [PubMed] [Google Scholar]
- 26.Edgerton M., Koshlukova S. E., Araujo M. W., Patel R. C., Dong J., Bruenn J. A. Salivary histatin 5 and human neutrophil defensin 1 kill Candida albicans via shared pathways. Antimicrob. Agents Chemother. 2000;44:3310–3316. doi: 10.1128/aac.44.12.3310-3316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aley S. B., Zimmerman M., Hetsko M., Selsted M. E., Gillin F. D. Killing of Giardia lamblia by cryptdins and cationic neutrophil peptides. Infect. Immun. 1994;62:5397–5403. doi: 10.1128/iai.62.12.5397-5403.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Yu W., He T., Yu J., Caffrey R. E., Dalmasso E. A., Fu S., Pham T., Mai J., Ho J. J., et al. Contribution of human α-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science. 2002;298:995–1000. doi: 10.1126/science.1076185. [DOI] [PubMed] [Google Scholar]
- 29.de Leeuw E., Burks S. R., Li X., Kao J. P., Lu W. Structure-dependent functional properties of human defensin 5. FEBS Lett. 2007;581:515–520. doi: 10.1016/j.febslet.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epand R. M., Vogel H. J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 31.Sitaram N., Nagaraj R. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochim. Biophys. Acta. 1999;1462:29–54. doi: 10.1016/s0005-2736(99)00199-6. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer R. I., Barton A., Daher K. A., Harwig S. S., Ganz T., Selsted M. E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Invest. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehrer R. I., Ganz T. Defensins of vertebrate animals. Curr. Opin. Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 34.Looft C., Paul S., Philipp U., Regenhard P., Kuiper H., Distl O., Chowdhary B. P., Leeb T. Sequence analysis of a 212 kb defensin gene cluster on ECA 27q17. Gene. 2006;376:192–198. doi: 10.1016/j.gene.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Schramm G., Bruchhaus I., Roeder T. A simple and reliable 5′-RACE approach. Nucleic Acids Res. 2000;28:e96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hristova K., Dempsey C. E., White S. H. Structure, location, and lipid perturbations of melittin at the membrane interface. Biophys. J. 2001;80:801–811. doi: 10.1016/S0006-3495(01)76059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waxman E., Rusinova E., Hasselbacher C. A., Schwartz G. P., Laws W. R., Ross J. B. Determination of the tryptophan:tyrosine ratio in proteins. Anal. Biochem. 1993;210:425–428. doi: 10.1006/abio.1993.1220. [DOI] [PubMed] [Google Scholar]
- 38.Chen G. C., Yang J. T. Two-point calibration of circular dichrometer with d-10-camphorsulfonic acid. Anal. Lett. 1977;10:1195–1207. [Google Scholar]
- 39.Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods. 1991;137:167–171. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 40.Leippe M., Ebel S., Schoenberger O. L., Horstmann R. D., Müller-Eberhard Pore-forming peptide of pathogenic Entamoeba histolytica. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7659–7663. doi: 10.1073/pnas.88.17.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 42.Soares T. A., Hunenberger P. H., Kastenholz M. A., Krautler V., Lenz T., Lins R. D., Oostenbrink C., van Gunsteren W. F. An improved nucleic acid parameter set for the GROMOS force field. J. Comput. Chem. 2005;26:725–737. doi: 10.1002/jcc.20193. [DOI] [PubMed] [Google Scholar]
- 43.Kraulis P. J. A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 44.Sreerama N., Woody R. W. Structural composition of βI- and βII-proteins. Protein Sci. 2003;12:384–388. doi: 10.1110/ps.0235003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann G., Müller P. A molecular model of membrane excitability. J. Supramol. Struct. 1974;2:538–557. doi: 10.1002/jss.400020504. [DOI] [PubMed] [Google Scholar]
- 46.Lynn D. J., Bradley D. G. Discovery of α-defensins in basal mammals. Dev. Comp. Immunol. 2007;31:963–967. doi: 10.1016/j.dci.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Raj P. A., Antonyraj K. J., Karunakaran T. Large-scale synthesis and functional elements for the antimicrobial activity of defensins. Biochem. J. 2000;347:633–641. [PMC free article] [PubMed] [Google Scholar]
- 48.Cullor J. S., Wood S., Smith W., Panico L., Selsted M. E. Bactericidal potency and mechanistic specificity of neutrophil defensins against bovine mastitis pathogens. Vet. Microbiol. 1991;29:49–58. doi: 10.1016/0378-1135(91)90109-s. [DOI] [PubMed] [Google Scholar]
- 49.Wimley W. C., Selsted M. E., White S. H. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]