Abstract
The tumour suppressor protein p53 regulates the expression of several genes that mediate cell cycle arrest, apoptosis, DNA repair and other cellular responses. Recently, we have shown that human transcriptional co-activator PC4 is a unique activator of p53 function. In the present study, we report that PC4 is a p53-inducible gene. Bioinformatics analysis reveals multiple p53-binding sites in the PC4 promoter. We have found that indeed p53 binds to all the identified sites in vitro and in vivo with varying affinities. p53 acts as an activator of PC4 transcription. Both PC4 mRNA and protein levels increase in response to stimuli that result in p53 induction. Furthermore, PC4 enhances p53 recruitment to the PC4 promoter. Our results thus establish the first report of a positively regulated feedback loop to control p53 function.
Keywords: chromatin immunoprecipitation, DNA damage, p53, PC4, promoter, transcription
Abbreviations: ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility-shift assay; GADD45, growth-arrest and DNA-damage-inducible protein 45; HEK-293 cells, human embryonic kidney cells; HEK-293T cells, HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40); Mdm2, murine double minute 2; RT, reverse transcriptase; TBP, TATA-box-binding protein
INTRODUCTION
The tumour suppressor protein p53 plays a pivotal role in maintenance of cellular homoeostasis. It is a sequence-specific transcription factor, binds as a tetramer and regulates the expression of several genes that are involved in diverse cellular processes like cell cycle arrest, apoptosis, DNA repair and angiogenesis [1–4]. The cellular level of p53 gets elevated by a number of stresses like DNA damage, hypoxia, oncogenes and nucleotide deprivation. p53 activation is mainly correlated with an increase or decrease in transcription of several (p53-responsive) genes that play important roles in these cellular processes. Activation and stabilization of p53 is regulated by specific post-translational modifications (phosphorylation, acetylation, methylation, ubiquitinylation and sumoylation). Apart from the post-translational modifications, p53-interacting proteins also play a major role in regulating p53 function. Among these, Mdm2 (murine double minute 2) and Pirh2 inhibit p53 activity in a regulated fashion [5–7]. Interestingly, the expression of these genes is in turn regulated by p53 [6–8]. p53 interacts with several proteins implicated in transcriptional regulation such as p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein], TBP (TATA-box-binding protein), TAF9 (TBP-associated factor 9), TAF6 (TBP-associated factor 6), Zac1, JMY (p300 junction mediating regulatory factor), HMGB1 (high-mobility group box 1), PC4 and hnRNPK (heterogeneous nuclear ribonucleoprotein K) [9–16]. TBP, by direct interaction with p53, stimulates binding to its cognate site and results in the enhancement of p53-dependent transcription [16]. Non-histone chromatin-associated protein HMGB1 acts as a unique activator of p53 by providing a bent DNA for p53 recruitment [13]. Recently, we have shown that multifunctional transcriptional co-activator PC4 activates p53 function [14]. PC4 is a highly abundant nuclear protein, which plays an important role in transcription, replication, DNA repair and cellular transformation [17–21]. PC4 is highly conserved from yeast to human, suggesting its importance in cellular function. Presumably, by stimulating activator-dependent transcription, PC4 plays a critical role in the maintenance of normal cellular growth. PC4 can inhibit AP2 (activator protein 2) self-repression in a ras-transformed cell line and may act as a putative tumour suppressor [20]. PC4 directly interacts with p53 in vitro and in vivo and enhances its DNA binding as well as its transcriptional activity. Although much attention has been focused on functional aspects of PC4, the regulation of PC4 gene expression has not been explored yet. Functional analysis of regulatory regions of the PC4 gene will help in understanding the role of PC4 in a cellular context. Here we show that p53 binds to PC4 promoter in vitro and in vivo thereby regulating PC4 gene expression. Furthermore, under DNA-damaging conditions, p53 gets recruited to the PC4 promoter and elevates the expression of PC4. Interestingly, PC4 also activates the binding of p53 to the PC4 promoter. Our results clearly show the positive autoregulation of p53 by activating the p53 activator PC4 expression.
MATERIALS AND METHODS
Bioinformatics analysis
The upstream sequence extraction homology searches were performed by BLAST search using the website http://www.ncbi.nlm.nih.gov. Alignments of PC4 protein sequences were done using the ClustalW program (http://www.ebi.ac.uk/clustalw). Target Explorer (http://www.trantor.bioc.columbia.edu) and MatInspector (http://www.genomatix.de) were used to search for p53-binding sites in the PC4 gene upstream sequence.
Plasmid construction
Genomic DNA used for cloning the upstream sequence of the PC4 gene was isolated from human blood using a commercial kit (Qiagen). The upstream sequences were cloned into pGL3-Basic vector (Promega) by using a PCR-based technique using specific primers at KpnI and XhoI enzyme cloning sites. The primer sets used for cloning: −1944 to +56 region of PC4 promoter are 5′-CGGGGTACCGGATCAAGGATTTTCGATTAG-3′ and 5′-CCGCTCGAGCGCTTCGCTCGGCTCTAGCAG-3′; −3916 to −1944 region are 5′-CGGGGTACCGGAGCTCGGAGAGCTCGGCTG-3′ and 5′-CCGCTCGAGGCGTTGCCCATTTTTAAAAGG-3′; −1000 to +1056 region are 5′-CGGGGTACCGATCCAAACACTCCTCCCTTTC-3′ and 5′-CCGCTCGAGGCCTGGGGGCCTAAGAATTTTTC-3′; −3000 to −1056 region are 5′-CGGGGTACCGTAATTTATGCCTGGAATCC-3′ and 5′-CCGCTCGAGCATATGGAGCAGTTAGACCTG-3′.
Purification of recombinant proteins
Recombinant N-terminal FLAG epitope-tagged human p53 and native PC4 were expressed and purified from Escherichia coli as described elsewhere [22,23].
EMSA (electrophoretic mobility-shift assay)
A single-stranded oligonucleotide (40-mer) corresponding to the putative p53-responsive sites or the mutated p53-responsive sites of the PC4 promoter was end-labelled with T4 polynucleotide kinase (Invitrogen). The radiolabelled strand was annealed with the complementary strand, and an EMSA was performed as described previously [13]. For the unlabelled oligonucleotide challenge experiments, the double-stranded unlabelled 40-mer oligonucleotide or GADD45 (growth-arrest and DNA-damage-inducible protein 45) promoter oligonucleotide (100-fold excess) was used. For supershift assays, the indicated amounts of antibodies were added into the reaction mixture prior to the EMSA reaction. The dissociation constants for p53 and the oligonucleotides were determined using Scatchard plot analysis as described previously [24].
Cell culture and transfection assay
H1299 (p53−/−), HCT116 (p53+/+), MCF7 (p53+/+), A549 (p53+/+), HEK-293 (human embryonic kidney cells) and HEK-293T [HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40)] (p53+/+) cells were maintained at 37 °C in DMEM (Dulbecco's modified Eagle medium) with 10% (v/v) FBS (Invitrogen). Prior to transfection, cells were seeded at 0.6×106 cells per 60 mm-diameter dishes and transfected with different plasmid constructs as indicated using Lipofectamine™ 2000 Plus (Invitrogen) according to the manufacturer's protocol. Luciferase and β-galactosidase activities were measured 24 h after the transfection with the luciferase assay and β-galactosidase assay systems according to the protocol provided by the manufacturer (Promega). The transient transfection assay was performed three times independently to monitor average relative luciferase activity.
Endogenous gene expression assay
H1299, HCT116, MCF7, A549 and HEK-293 cells were treated with cisplatin (15 μg/ml) for 12 h and doxorubicin (1–2 μg/ml) for 24 h. Total RNA was isolated from the treated cells using TRIzol® reagent (Invitrogen) and cDNA was made using oligo-dT (28-mer) and MMLV (Moloney-murine-leukaemia virus) RT (reverse transcriptase) (Invitrogen). This cDNA was used for real-time PCR analysis using iQ™ SYBR Green Supermix (Bio-Rad) with gene-specific primers. Whole cell lysates were made and immunoblotting was performed using the indicated antibodies and β-actin as loading control.
ChIP (chromatin immunoprecipitation) assay
HEK-293 cells, treated with or without doxorubicin (for 24 h), were used for ChIP assay. ChIP assay was performed as described elsewhere [25]. Briefly, HEK-293 cells were subjected to doxorubicin treatment (1 μg/ml) for 24 h. Cross-linking was performed with 1% formaldehyde followed by cell lysis in SDS lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris/HCl, pH 8). After sonication of the chromatin (six times for 10 s at power setting 91%), cold dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris/HCl, pH 8, and 167 mM NaCl) was added along with preblocked Protein G–Sepharose (Amersham Biosciences) and monoclonal anti-p53 antibody DO1 (Oncogene) and kept for overnight binding. The sonicated samples were precleared prior to immunoprecipitation. Beads were washed with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8, and 150 mM NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8, and 500 mM NaCl), LiCl buffer [250 mM LiCl, 1% NP40 (Nonidet P40), 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris/HCl, pH 8] and TE (10 mM Tris/HCl, pH 8, and 1 mM EDTA) consecutively. To the washed beads, elution buffer (0.2% SDS and 100 mM NaHCO3) was added along with 200 mM NaCl and kept overnight at 65 °C. Proteinase K (0.1 mg/ml; Sigma) and 0.04 mg/ml of RNase A (Sigma) were added and incubated subsequently for 2 h at 55 °C. The immunoprecipitated samples were deproteinized, ethanol-precipitated and used for real-time PCR analysis. Primer sets corresponding to the p53-binding sites used in RT–PCR analysis are: 5′-TTCCTCTACCTGAGGCAGG-3′ and 5′-ATAGCTAAAAGATTATTTG-3′ for p53.1; 5′-TTGTATATGGATTCCTC-3′ and 5′-TGCGTTTGTTCAATAAATG-3′ for p53.2; 5′-TGAGGTGGGTGAAGTGGGG-3′ and 5′-GGACCGCGTCTCTTCCTAAATG-3′ for p53.3; 5′-GCTATCTCCTACAGGAAGG-3′ and 5′-AGTCGAACACCCTCTTGGTCG-3′ for p53.4; and 5′-CAGGCTGTGGCTCTGATTGG-3′ and 5′-TTCAGAGTAACAGGCTAAGG-3′ for p21RE1.
RESULTS
Genomic organization of the PC4 gene in humans
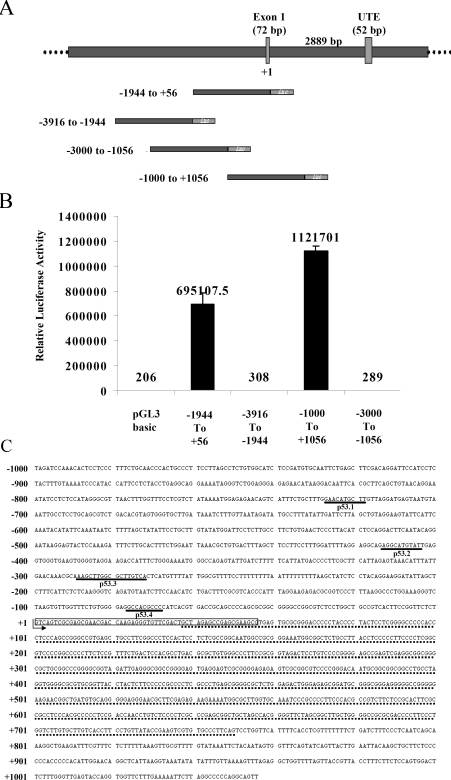
The multifunctional transcriptional co-activator PC4 is highly conserved across the species. Homologues of PC4 are identified in mouse, rat, yeast (SUB1), Arabidopsis (hypothetical), Caenorhabditis (hypothetical) and Drosophila. Multiple alignment of these homologues using the ClustalW program, clearly indicates the strong identity (‘*’) and similarity (‘.’) among them (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/406/bj4060437add.htm). The size and percent identity of the homologues are given in Supplementary Table 1 (http://www.BiochemJ.org/bj/406/bj4060437add.htm). The BLAST search of the human genome of NCBI (National Center for Biotechnology Information) and NIH (National Institutes of Health) database yielded five different gene sequences, highly similar to the PC4 gene in different human chromosomes (see Supplementary Figures 2A and 2B at http://www.BiochemJ.org/bj/406/bj4060437add.htm). Analysis of these sequences revealed that PC4 genes present in the chromosomes 14 and 8 have a single exon. The sequence in chromosome 5 has five exons including one 5′-UTE (untranslated element) and the chromosome 16 contains two partial sequences similar to the PC4 gene (Supplementary Figures 2A and 2B). However, the sequences in chromosomes 8, 14 and 16 are presumably pseudogenes (http://www.pseudogene.org). Therefore promoter analysis was carried out with the candidate gene present in chromosome 5. We have cloned the upstream region of PC4 in chromosome 5 into a promoter-less luciferase reporter plasmid (pGL3-Basic; Promega) to evaluate the promoter activity. The different fragments of 2 kb upstream regions (Figure 1A) were cloned and the promoter activity of each construct was measured by transfecting these constructs in HEK-293T cells followed by luciferase assay (Figure 1B). The construct −1000 to +1056, which contains the 713 bp GC-rich region (Figure 1C), showed the highest promoter activity (Figure 1B). In order to verify the web-based information, the upstream region of the PC4 pseudogene in chromosome 8 was also cloned and tested for promoter activity. As it was predicted these constructs did not show any promoter activity (results not shown).
Figure 1. Analysis of the PC4 genomic locus.
(A) The upstream sequence of PC4 exon I was extracted from Entrez (http://www.ncbi.nlm.nih.gov) and different promoter constructs were made in pGL3 Basic vector (Promega) with a luciferase reporter. (B) These constructs (200 ng) and an internal control CMV-β-Gal were used for transfection of 0.5 million cells with Lipofectamine™ 2000 (Invitrogen). Luciferase activity was assayed with a luciferase kit (Promega) 24 h after transfection. The assay was repeated three times in duplicate and the average is represented in the Figure. The average luciferase counts are presented on each bar. (C) The promoter region 2056 bp (–1000 to +1056) showed five p53-binding sites (two overlapping) and a GC-rich region (underlined by a dotted line). The p53-binding sites are underlined and labelled.
p53 binds to the PC4 promoter in vitro and in vivo
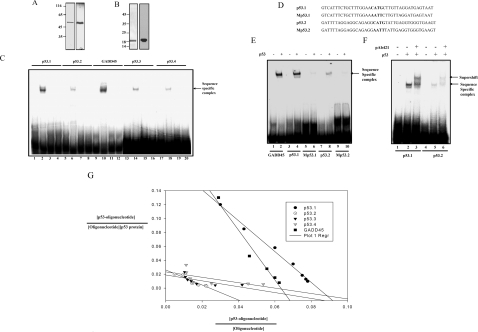
Promoter analysis of PC4 did not show any TATA-box sequence but it possesses binding sites for a large number of transcription factors. However, the bioinformatics analysis of the promoter construct from bases −1000 to +1050 using MatInspector and Target explorer suggests the presence of putative p53-binding sites. This region contains five p53-binding half-sites, out of which two are found to be overlapping (Figures 1A and 1C). To analyse p53 binding to these sites, recombinant bacterially expressed N-terminal FLAG epitope-tagged human wild-type p53 was purified (Figure 2A) and gel-shift assays were performed. Radiolabelled 40-mer-oligonucleotide sequences derived from putative binding sites named as p53.1, p53.2, p53.3, p53.4 and p53 consensus binding site from the p53-responsive gene GADD45 (positive control) were used. p53 could form a sequence-specific complex with all of the identified sites in the PC4 promoter (Figure 2C, lanes 2, 6, 14 and 18). Formation of a sequence-specific complex was competed out when 100-fold excess of unlabelled consensus oligonucleotide (GADD45) (Figure 2C, lanes 4, 8, 16 and 20) or the same oligonucleotide (Figure 2C, lanes 3, 7, 15 and 19) was added in the same reaction. In order to ensure the site-specific binding of p53 (to the PC4 promoter), the binding sites in p53.1 and p53.2 were mutated (Figure 2D). Significantly, these mutations reduced p53 binding to the oligonucleotides p53.1 and p53.2 (Figure 2E, lane 4 compared with lane 6 and lane 8 compared with lane 10). Furthermore, 100-fold excess addition of unlabelled mutated oligonucleotides could not inhibit the formation of a p53 oligonucleotide complex in unlabelled competition experiments (results not shown). The presence of p53 in the protein–DNA complex was further confirmed by supershift assay using p53 monoclonal antibody pAb421 (Figure 2F, lanes 3 and 6). Taken together, these results suggest that p53 forms a sequence-specific complex with the identified binding sites in the PC4 promoter in vitro. To analyse the strength of p53 binding to the different PC4 promoter sites, dissociation constants (Kd) for each site were determined. Oligonucleotides were incubated with different concentrations of p53 (see Supplementary Figure 3 at http://www.BiochemJ.org/bj/406/bj4060437add.htm) (see the Materials and methods section) and the sequence-specific complexes were quantified using phosphoimager analysis. The data were plotted according to Scatchard and the Kd values were calculated (Figure 2G and Table 1). Results show that the affinities for p53 are in the order of GADD45>p53.1>p53.2>p53.3>p53.4. Interestingly, the binding affinity of the p53.1 site was comparable with that of the canonical binding site of p53 in the GADD45 promoter.
Figure 2. p53 binds to the PC4 promoter.
Purification of recombinant proteins: (A) 100 ng of recombinant FLAG-tagged p53 was analysed by SDS/10% PAGE (left panel) and authenticity of the p53 was analysed by Western blotting using anti-p53 monoclonal antibody PAb421 (right panel). (B) Recombinant PC4 (300 ng) was analysed by SDS/15% PAGE (left panel), and authenticity of the PC4 was analysed by Western blotting using anti-PC4 polyclonal antibody (right panel). (C) EMSAs were performed using radiolabelled p53.1, p53.2, GADD45, p53.3 and p53.4 oligonucleotides, incubated either with purified recombinant p53 protein (lanes 2, 6, 10, 14 and 18) alone or in combination with 100-fold excess of unlabelled oligonucleotide (lanes 3, 7, 11, 15 and 19) or 100-fold excess of unlabelled p53 consensus (GADD45) oligonucleotide (lanes 4, 8, 12, 16 and 20). Lanes 1, 5, 9, 13 and 17 represent the 32P-labelled p53.1, p53.2, GADD45, p53.3 and p53.4 oligonucleotides alone. (D) Sequences of oligonucleotides p53.1 and p53.2 containing p53-binding sites and Mp53.1 and Mp53.2 containing mutated p53-binding sites. (E) EMSAs were performed using radiolabelled GADD45, p53.1, Mp53.1, p53.2 and Mp53.2 oligonucleotides incubated with purified recombinant p53 protein (lanes 2, 4, 6, 8 and 10). Lanes 1, 3, 5, 7 and 9 represent the 32P-labelled GADD45, p53.1, Mp53.1, p53.2 and Mp53.2 respectively. (F) Supershift assays were performed by incubation of p53 with p53 antibody prior to addition of oligonucleotides p53.1 and p53.2 (lanes 3 and 6). Lanes 2 and 5 represent oligonucleotides p53.1 and p53.2 incubated with p53 alone and lanes 1 and 4 represent 32P-labelled p53.1 and p53.2 oligonucleotides alone. (G) Determination of Kd of p53 binding to p53.1, p53.2, p53.3, p53.4 and GADD45. EMSAs were performed using radiolabelled p53.1, p53.2, p53.3, p53.4 and GADD45 either alone or with increasing concentrations (5–162 ng) of recombinant purified p53 protein. The binding constant (Kd) of p53 binding to each oligonucleotide was determined according to Scatchard plot (see Table 1).
Table 1. Oligonucleotides used in EMSA and p53 binding constants.
Sequence and binding constant values (Kd) of each p53-binding site in the PC4 promoter, along with the GADD45 p53-binding site are shown.
| Primer | Sequence | Kd |
|---|---|---|
| p53.1 | GTCATTTCTGCTTTGGAACATGCTTGTTAGGATGAGTAAT | 4.5 nM |
| p53.2 | GATTTTAGGAGGCAGAGGCATGTATTGAGGTGGGTGAAGT | 9.3 nM |
| p53.3 | TGAACAAACGCAAAGCTTGGCGCTTGTCACTCATGTTTTAT | 17.8 nM |
| p53.4 | TGGTTTCTGTGGGGAGGCCACGCCCCATCACGTGACCGCA | 22.0 nM |
| GADD | AATTCTCGAGCAGAACATGTCTAAGCATGCTGGGCTCGAG | 4.1 nM |
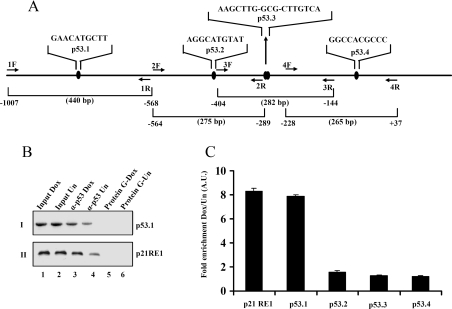
In order to find out the physiological relevance of these results, we further investigated whether p53 binds to the PC4 promoter in vivo. ChIP assays were performed using p53 antibody followed by PCR analysis with site-specific primers for different p53-binding sites (Figure 3A) in the PC4 promoter. As expected, we observed that there is an enrichment of p53 binding to the p21 RE1 site upon doxorubicin treatment (Figure 3B, panel II, lane 3 compared with lane 4). Among all the p53-binding sites in the PC4 promoter, the fold enrichment of p53 binding was most pronounced in the case of the p53.1 site (Figure 3B, panel I, lane 3 compared with lane 4). Further quantitative estimation of the fold enrichment of p53 recruitment upon doxorubicin treatment was performed by real-time PCR analysis (Figure 3C). The presence of basal levels of p53 was found on all identified binding sites of the PC4 promoter in untreated cells. However, the association of p53 is enhanced to ∼8-fold only at the p53.1 site of the PC4 promoter, similar to the p21 promoter, upon doxorubicin treatment (Figure 3C). These results clearly indicate that p53 is recruited to the PC4 promoter in vivo.
Figure 3. p53 binds to the PC4 promoter in vivo.
HEK-293 cells were treated with doxorubicin for 24 h and ChIP assays were performed using p53 antibody. (A) Sequences of the p53-binding sites in the PC4 promoter. p53.1 F and R, p53.2 F and R, p53.3 F and R, and p53.4 F and R represent the primer sets used for PCR amplification of the respective sites. The amplicon sizes and primer sets are labelled. (B) ChIP assay was performed with or without anti-p53 antibody. The final DNA extractions were amplified using specific primers for p53-binding sites in PC4 (I) and p21 (II) promoters. (C) The fold enrichment of p53 recruitment upon doxorubicin treatment in p53-binding sites of the PC4 promoter and p21 promoter.
p53 enhances PC4 gene expression in vivo
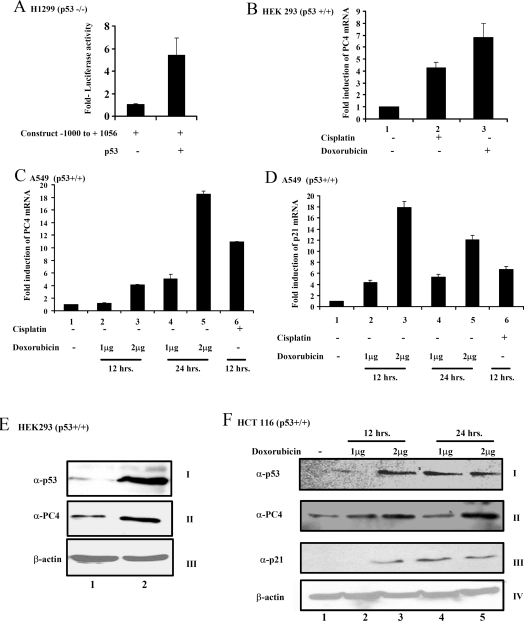
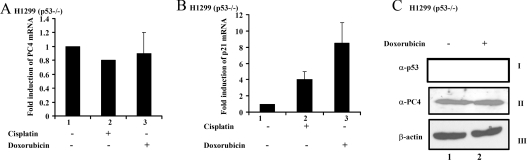
After establishing that the putative sites for p53 binding in the PC4 promoter are indeed physiologically active, we tested the activity of the PC4 promoter in the p53-negative background. H1299 cells (p53−/−) were transfected with the PC4 promoter construct either alone or in combination with the mammalian expression construct for p53. The transfection efficiency was normalized with β-galactosidase activity. Significantly, a p53-dependent 5-fold induction of PC4 promoter activity could be observed in the H1299 cells (Figure 4A), suggesting that p53 can act as a transcriptional activator of PC4 gene expression. To assess p53-mediated regulation of PC4 endogenous gene expression, we examined PC4 mRNA and protein levels following treatment with p53-inducing agents. The wild-type p53-containing cell lines, A549, HCT116, HEK-293 and MCF7, were used for this purpose. In agreement with the above-mentioned results, upon treatment with doxorubicin or cisplatin, an increased level of PC4 mRNA was detected by real-time PCR analysis [Figures 4B (lane 1 compared with lanes 2 and 3), 4C (lane 1 compared with lanes 2–6) and Supplementary Figures 4A, panel I (lane 1 compared with lanes 2–6) and 4B, panel I (lane 1 compared with lanes 2 and 3) at http://www.BiochemJ.org/bj/406/bj4060437add.htm], which is comparable with the p21 mRNA levels [Figure 4D (lane 1 compared with lanes 2–6)]. The PC4 protein levels were also increased upon doxorubicin treatment in p53-positive cells (panel II in Figures 4E and 4F and Supplementary Figures 4D and 4E) with a concomitant increase in p53 levels (panel I in Figures 4E and 4F and Supplementary Figures 4D and 4E) and p21 levels (panel III in Figure 4F and Supplementary Figures 4D and 4E) as shown by Western blotting. In contrast, PC4 expression both at mRNA and protein levels did not show any change in p53 null cell line (H1299) upon treatment with doxorubicin/cisplatin [Figures 5A (lane 1 compared with lanes 2 and 3) and 5C, panel II (lane 1 compared with lanes 2 and 3), and Supplementary Figure 4C, panel I (lane 1 compared with lanes 2 and 3)]. However, p21 overexpression is observed [Figure 5B (lane 1 compared with lanes 2 and 3)]. These results show that PC4 overexpresses upon genotoxic insult and the elevation of gene expression is p53-dependent.
Figure 4. PC4 overexpresses upon genotoxic insult.
(A) H1299 (p53−/−) cells were transiently transfected with PC4 promoter construct −1000 to +1056 (500 ng) alone or along with mammalian expression construct of p53 (2 μg). After 24 h of transfection the luciferase activity was measured and normalized with β-galactosidase activity. Each experiment was repeated three times and mean and S.D. is represented in the Figure. (B) HEK-293 (p53+/+) cells and (C, D) A549 (p53+/+) cells were treated with cisplatin for 12 h or 1 or 2 μg/ml of doxorubicin for 12 or 24 h. RNA was isolated and real-time PCR analysis was performed using specific primers for PC4 and p21 with β-actin as a control. Fold induction of PC4 and p21 mRNA levels is represented. (E) HEK-293 (p53+/+) cells were treated with doxorubicin for 24 h. Whole cell lysates were made from the treated/untreated cells and immunoblotting was performed for PC4, p53 and β-actin. (F) HCT116 (p53+/+) cells were treated with 1 and 2 μg/ml doxorubicin for 12 and 24 h. Whole cell lysates were made from the treated/untreated cells and immunoblotting was performed for PC4, p53, p21 and β-actin.
Figure 5. p53-dependent overexpression of PC4.
(A, B) H1299 (p53−/−) cells were treated with cisplatin for 12 h and doxorubicin for 24 h. RNA was isolated and real-time PCR analysis was performed using specific primers for PC4 and p21 with β-actin as a control. (C) H1299 (p53−/−) cells were treated with doxorubicin for 24 h. Whole cell lysates were made from the treated/untreated cells and immunoblotting was performed for PC4, p53 and β-actin.
Activation of p53 recruitment by PC4 to the PC4 promoter
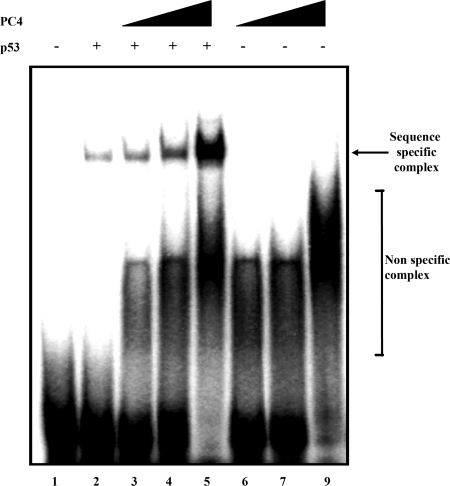
We have recently shown that PC4 enhances the DNA binding ability of p53 to its cognate binding sites [14]. In vitro and in vivo studies show that p53 binds to the PC4 promoter, which tempted us to find out whether PC4 enhances the binding of p53 to the PC4 promoter. Recombinant bacterially expressed PC4 was purified and authenticity was confirmed by immunoblotting with PC4 antibodies (Figure 2B). A 40-mer-oligonucleotide-containing p53.1 site of the PC4 promoter was used in the EMSA assay for this purpose. Interestingly, we found that activation of p53 binding by PC4 to its own promoter was indeed in a dose-dependent manner (Figure 6, compare lane 2 with lanes 3, 4 and 5). These results establish that the p53 activator, PC4, is regulated by p53 and the autoregulatory loop acts positively in recruiting p53 to PC4 promoter.
Figure 6. Activation of p53 recruitment by PC4 to its own promoter.
32P-labelled p53.1 oligonucleotide (3 ng) containing a p53-binding site was incubated with 50 ng of p53 either in the absence of PC4 (lane 2) or in the presence of 50 ng (lane 3), 100 ng (lane 4) or 200 ng (lane 5) of PC4. The radiolabelled oligonucleotide was also incubated with 50 ng (lane 6), 100 ng (lane 7) or 200 ng (lane 8) of PC4 alone. Lane 1 contains the 32P-labelled oligonucleotide.
DISCUSSION
p53 represents the central integrator of signals emanating from cellular DNA damage and resulting in cell cycle arrest, DNA repair and cell death. Mechanistically p53 acts as a transcriptional activator (often also as a repressor) and regulates the expression of genes involved in these cellular processes. Several proteins on the other hand play a pivotal role in p53-mediated transcriptional regulation through their direct interaction with p53. Multifunctional human transcriptional co-activator PC4 is one such example. Significantly, our results establish that PC4 is a downstream target of p53. This is evidenced by (i) presence of multiple binding sites in the promoter of the gene, (ii) increase in the in vivo association of p53 to the PC4 promoter upon genotoxic insult and (iii) p53-dependent activation of endogenous PC4 gene expression.
The multifunctional transcriptional co-activator PC4 is a highly abundant nuclear protein, which plays an important role in transcription, replication, DNA repair, cellular transformation and also chromatin organization [17–21]. Here we elucidate the regulatory elements involved in PC4 gene expression, which would be essential to understand PC4 function in a global perspective. In humans, a functional PC4 gene is present on chromosome 5. We have cloned and sequenced the upstream region of the PC4 gene and demonstrated its promoter activity (Figure 1B). Examination of the PC4 promoter revealed several potential binding sites for transcription factors, which are linked to distinct signalling pathways. As a remarkable property of the PC4 promoter we have identified multiple binding sites for tumour suppressor p53. Presence of multiple p53-binding sites is a characteristic feature of several physiologically important p53-responsive genes such as p21, Cyclin G and p22/PRG1 [26–28]. Our results suggest that p53 does indeed bind to the identified sites of the PC4 promoter in vitro and in vivo and plays a key role in the regulation of PC4 gene expression. The binding affinity of p53 for the p53.1 site was found to be comparable with the binding affinity towards the p53-binding site from the human GADD45 (a well-known p53-responsive gene) promoter, emphasizing the role of p53 in the transcriptional regulation of PC4. ChIP assays after genotoxic insult revealed predominant association of p53 with the p53.1 site of the PC4 promoter in HEK-293 cells (Figure 3). The fold enrichment of p53 binding to the PC4 promoter at the p53.1 site was found to be similar to RE1 site of p21 promoter, suggesting the physiological significance of p53 recruitment to the PC4 promoter in vivo. However, p53 recruitment to other sites in the PC4 promoter is not enriched upon genotoxic insult. Possibly these binding sites may respond to some other cellular signals.
The functional significance of p53 binding to the PC4 promoter was verified upon genotoxic insult to different cell lines containing wild-type p53 (A549, HCT116, HEK-293 and MCF7). Interestingly, the elevated p53 protein levels upon genotoxic insult resulted in a concomitant increase in PC4 mRNA and protein levels (Figure 4). In contrast, p53 null cells (H1299) did not show PC4 induction upon genotoxic insult, suggesting that the DNA damage-induced activation of PC4 expression is p53-dependent (Figure 5). PC4, through its direct interaction with p53, activates p53-mediated DNA binding and thereby induces p53-responsive gene expression and apoptosis as shown in our previous work [14]. Interestingly, overexpression of PC4 causes apoptosis [14]. Therefore the up-regulation of PC4 gene expression upon genotoxic insult presumably plays a significant role in p53-dependent pathways such as apoptosis, cell cycle arrest and DNA repair. Furthermore, enhancement of p53 recruitment by PC4 to the PC4 promoter and also activation of p53 function by PC4 suggest that overexpression of PC4 may contribute to p53-dependent function in DNA-damaged cells. PC4 plays an important role in DNA repair by preventing mutagenesis and killing by oxidative DNA damage [19]. Absence of the PC4 yeast homologue Sub1 renders the cells to undergo spontaneous and peroxide-induced hypermutability, suggesting the importance of PC4 overexpression under DNA-damaging conditions and the possible involvement of PC4 in double-strand break repair.
p53 has a large number of transcriptional targets, which include several p53-regulatory protein genes like Mdm2, Pirh2 and COP1 [5–7]. Significantly, all the three genes inhibit the p53 activity in different pathways. Therefore the present finding that the activator of p53, PC4, is also a transcription target of the tumour suppressor attributes a new function to p53. p53 gets mutated in several human cancers most of which are impaired in the DNA binding ability of p53. Since PC4 is a multifunctional protein and its expression is regulated by p53, it would be interesting to find out the alteration of PC4 function in these p53 mutation backgrounds, and its functional significance.
Online data
Acknowledgments
We thank the Indian Council of Medical Research, Government of India and Jawaharlal Nehru Centre for Advanced Scientific Research for financial support. We thank Dr Ghanshyam Swarup (Centre for Cellular and Molecular Biology, Hyderabad, India) for providing us valuable reagents. K. B. is a CSIR (Council of Scientific and Industrial Research) senior research fellow.
References
- 1.El-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 2.Miyashita T., Reed J. C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 3.Kastan M. B., Zhan Q., El-Deiry W. S., Carrier F., Jacks T., Walsh W. V., Plunkett B. S., Vogelstein B., Fornace A. J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 4.Dameron K. M., Volpert O. V., Tainsky M. A., Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 5.Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 6.Dornan D., Wertz I., Shimizu H., Arnott D., Frantz D. G., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 7.Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 8.Juven T., Barak Y., Zaruberman A., George D. L., Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;12:3411–3416. [PubMed] [Google Scholar]
- 9.Lill N. L., Grossman S. R., Ginsberg D., De Caprio J., Livingston D. M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 10.Ko L. J., Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 11.Huang S. M., Schonthal A. H., Stallcup M. R. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene. 2001;12:2134–2143. doi: 10.1038/sj.onc.1204298. [DOI] [PubMed] [Google Scholar]
- 12.Shikama N. C., Lee C. W., France S., Delavaine L., Lyon J., Demonacus K. M., La Thangue N. B. A novel cofactor for p300 that regulates the p53 response. Mol. Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman L., Moorthy N. C., Murthy K. G., Manley J. L., Bustin M., Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S., Kumar B. R., Kundu T. K. General transcriptional coactivator PC4 activates p53 function. Mol. Cell. Biol. 2004;24:2052–2062. doi: 10.1128/MCB.24.5.2052-2062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moumen A., Masterson P., O'Connor M. J., Jackson S. P. HnRNP: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Chen X., Farmer G., Zhu H., Prywes R., Prives C. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- 17.Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 18.Pan Z. Q., Ge H., Amin A. A., Hurwitz J. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J. Biol. Chem. 1996;271:22111–22116. doi: 10.1074/jbc.271.36.22111. [DOI] [PubMed] [Google Scholar]
- 19.Wang J. Y., Sarker A. H., Cooper P. K., Volkert M. R. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol. Cell. Biol. 2004;24:6084–6093. doi: 10.1128/MCB.24.13.6084-6093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannan P., Tainsky M. A. Coactivator PC4 mediates AP-2 transcriptional activity and suppresses ras-induced transformation dependent on AP-2 transcriptional interference. Mol. Cell. Biol. 1999;19:899–908. doi: 10.1128/mcb.19.1.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das C., Hizume K., Batta K., Kumar B. R., Gadad S. S., Ganguly S., Lorain S., Verreault A., Sadhale P. P., Takeyasu K., Kundu T. K. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell. Biol. 2006;26:8303–8315. doi: 10.1128/MCB.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu W., Roeder R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 23.Ge H., Roeder R. G. Purification, cloning, and characterization of a human coactivator, PC4 that mediates transcriptional activation of class II genes. Cell. 1994;78:518–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 24.Miao F., Bouziane M, O'Connor T. R. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Macleod K. F., Sherry N., Hannon G., Beach D., Tokino T., Kinzler K., Vogelstein B., Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 27.Zauberman A., Lupo A., Oren M. Identification of p53 target genes through immune selection of genomic DNA: the cyclin G gene contains two distinct p53 binding sites. Oncogene. 1995;10:2361–2366. [PubMed] [Google Scholar]
- 28.Schafer H., Trauzold A., Sebens T., Deppert W., Folsch U. R., Schmidt W. E. The proliferation-associated early response gene p22/PRG1 is a novel p53 target gene. Oncogene. 1998;16:2479–2487. doi: 10.1038/sj.onc.1201788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.