Abstract
The involvement of CK1 (casein kinase 1) δ in the regulation of multiple cellular processes implies a tight regulation of its activity on many different levels. At the protein level, reversible phosphorylation plays an important role in modulating the activity of CK1δ. In the present study, we show that PKA (cAMP-dependent protein kinase), Akt (protein kinase B), CLK2 (CDC-like kinase 2) and PKC (protein kinase C) α all phosphorylate CK1δ. PKA was identified as the major cellular CK1δCK (CK1δ C-terminal-targeted protein kinase) for the phosphorylation of CK1δ in vitro and in vivo. This was implied by the following evidence: PKA was detectable in the CK1δCK peak fraction of fractionated MiaPaCa-2 cell extracts, PKA shared nearly identical kinetic properties with those of CK1δCK, and both PKA and CK1δCK phosphorylated CK1δ at Ser370 in vitro. Furthermore, phosphorylation of CK1δ by PKA decreased substrate phosphorylation of CK1δ in vitro. Mutation of Ser370 to alanine increased the phosphorylation affinity of CK1δ for β-casein and the GST (gluthatione S-transferase)–p53 1–64 fusion protein in vitro and enhanced the formation of an ectopic dorsal axis during Xenopus laevis development. Anchoring of PKA and CK1δ to centrosomes was mediated by AKAP (A-kinase-anchoring protein) 450. Interestingly, pre-incubation of MiaPaCa-2 cells with the synthetic peptide St-Ht31, which prevents binding between AKAP450 and the regulatory subunit RII of PKA, resulted in a 6-fold increase in the activity of CK1δ. In summary, we conclude that PKA phosphorylates CK1δ, predominantly at Ser370 in vitro and in vivo, and that site-specific phosphorylation of CK1δ by PKA plays an important role in modulating CK1δ-dependent processes.
Keywords: A-kinase-anchoring protein (AKAP), cAMP-dependent protein kinase (PKA), casein kinase 1 δ (CK1δ), CDC-like kinase 2 (CLK2), protein kinase B (Akt), protein kinase Cα (PKCα)
Abbreviations: AKAP, A-kinase-anchoring protein; CK1, casein kinase 1; CK1δCK, CK1δ C-terminal-targeted protein kinase; CLK2, CDC-like kinase 2; DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; GST, gluthatione S-transferase; HA, haemagglutinin; HRP, horseradish peroxidase; NP40, Nonidet P40; PKA, cAMP-dependent protein kinase; PKC, protein kinase C; PP, protein phosphatase
INTRODUCTION
The CK1 (casein kinase 1) group is unique within the superfamily of serine/threonine-specific kinases found in all eukaryotic organisms. So far, seven mammalian CK1 isoforms (α, β, γ1, γ2, γ3, δ and ϵ) and their splice variants have been characterized. They are all highly conserved within their kinase domains, but differ significantly in length and primary structure of their N-terminal and C-terminal regulatory domains (reviewed in [1]). CK1 isoforms can phosphorylate a wide range of substrates bearing either a canonical or a non-canonical consensus sequence [2–7]. As a result, they can influence the activity of numerous key regulatory proteins. Processes modulated by CK1 include cell cycle progression and cytokinesis [8], DNA repair and recombination [9–12], chromosome segregation, microtubule dynamics, kinetechore- and centrosome-specific functions [13–16] and apoptosis [17–20]. Consequently, the interest in developing inhibitors to specifically target CK1 family members has increased significantly in recent years.
Several mechanisms have been identified that modulate the activity of CK1 in vitro and in vivo. Alternative splicing represents a mechanism that influences substrate binding, turnover and subcellular localization of certain CK1 isoforms [1]. In addition, interaction with cellular proteins influences the subcellular localization of CK1 [21–23] and can inhibit its activity [24]. It has been reported that the subcellular localization of CK1δ is an important factor for the regulation of its kinase–substrate interactions and depends on the enzymatic activity of the kinase [15]. Furthermore, autophosphorylation provides an additional mechanism to down-regulate CK1 activity [25–27]. However, mechanisms exist to overcome the inhibitory effect of autophosphorylation [28]. Other processes that can regulate CK1 activity include stimulation of cells with insulin, viral transformation, treatment of cells with topoisomerase inhibitors or with γ-irradiation, all resulting in elevated CK1 activity [1]. The increase of CK1ϵ kinase activity by Wnt-induced dephosphorylation of CK1ϵ autophosphorylation sites, probably performed by PP (protein phosphatase) 2A, was shown in vivo [29]. Similarly, in neostriatal neurons, the metabotrophic glutamate receptors activate CK1ϵ by PP2B [30–32].
Differences in the expression and activity levels of CK1δ have been detected in organs of young adult Balb/c mice [33]. It has been suggested that the differences observed could be the result of alterations in the degree of autophosphorylation or to changes in the activity of cellular kinases which are able to phosphorylate CK1δ. However, the intramolecular autophosphorylation status of CK1δ makes it difficult to identify such cellular kinases. In the present study, GST (gluthatione S-transferase)- tagged CK1δ fusion proteins were used as substrates because they do not exhibit any kinase activity, and single fractions of fractionated MiaPaCa-2 cell extracts were used as the source of cellular kinases. We have identified PKA (cAMP-dependent protein kinase) as the main cellular kinase that phosphorylates CK1δ, predominantly at Ser370. In addition, we have demonstrated that Akt (protein kinase B), CLK2 (CDC-like kinase 2) and PKC (protein kinase C) α also phosphorylate CK1δ at Ser370, as well as at other sites within amino acids 305–375 in vitro. Moreover, phosphorylation of CK1δ by PKA negatively influenced the substrate phosphorylation efficiency of CK1δ. Mutation of Ser370 to alanine enhanced the ability of CK1δ to phosphorylate β-casein and GST–p53 1–64 in vitro and increased the formation of an ectopic dorsal axis during Xenopus development. Inhibition of the interaction of PKA with AKAPs (A-kinase-anchoring proteins) by the synthetic St-Ht31 inhibitory peptide led to a significant up-regulation of the activity of CK1δ in MiaPaCa-2 cells. Our results suggest that PKA has an important regulatory effect on the activity of CK1δ.
MATERIALS AND METHODS
Construction of plasmids
Various CK1δ fragments were amplified by PCR using wild-type rat CK1δ (GenBank® accession number NM_139060) as a template, and the primers used are listed in Table 1.
Table 1. Sequences of CK1δ-specific primers.
| Primer | Sequence |
|---|---|
| CK1δ1 | 5′-CGAATTCCATGGAGAGCAAGATCTACAAAATGAT-3′ |
| CK1δ2 | 5′-ACTCGAGTCGACAGCATCATCTGCAGCC-3′ |
| CK1δ3 | 5′-CGAATTCCATGGACTTTGGGCTGGCCAAG-3′ |
| CK1δ4 | 5′-ACTCGAGTCGACGTAGGTGGTACGTCGTG-3′ |
| CK1δ5 | 5′-CGAATTCCATGGAGCTGAGGGTCGGGAAT-3′ |
| CK1δ6 | 5′-ACTCGAGTCGACTCAACGGTGCAGCCGCA-3′ |
| CK1δ7 | 5′-ACTCGAGTCGACTGAGGTAGGGGTAAGG-3′ |
| CK1δ8 | 5′-CGAATTCCATGGAGCGGGAACGCCG-3′ |
| CK1δ9 | 5′-CCATGGCTTCCGGCCGTCTGCGGGGAACC-3′ |
| CK1δ10 | 5′-ACTCGAGTCGACTCAAGGAGAGGTGTTGGC-3′ |
| CK1δ11 | 5′-CCATGGCCCGGAATCCAGCCACTCGTGGC-3′ |
| CK1δ12 | 5′-ACTCGAGTCGACTCAATGTGAGGTAGGGGT-3′ |
| CK1δ13 | 5′-ACTCGAGTCGACTCATGGGGGAGCCACTTC-3′ |
| CK1δ14 | 5′-CCATGGCTCCCCCAACGCCCCTTACCCCT-3′ |
| CK1δ15 | 5′-CCATGGCCAACACCTCTCCTAGACCCGTC-3′ |
| CK1δ16 | 5′-ACTCGAGTCGACTCAACGGTGCAGCCGCATAGCCAC-3′ |
| CK1δ17 | 5′-CCATGGCCCGGAATCCAGCCGCTCGTGGC-3′ |
| CK1δ18 | 5′-CCATGGCTCGTGGCCTCCCTTCTGCAGCT-3′ |
| CK1δ19 | 5′-CCATGGCCGGCCGTCTGCGGGGAACCCAG-3′ |
| CK1δ20 | 5′-CGAGAACGGAAAGTGGCTATGCGGCTGCACCGTGGG-3′ |
| CK1δ21 | 5′-GCTCTTGCCTTTCACCGATACGCCGACGTGGCACCC-3′ |
| CK1δ22 | 5′-GATATCATGTACCCATACGATGTTCCAGATTACGCTCTTCATATGGCGATGGAGCTGAGG-3′ |
| CK1δ23 | 5′-AGATCTTCAGTAGGTGGTACG-3′ |
| CK1δ24 | 5′-GGTACCATGGAGGAGCAGAAGCTG-3′ |
| CK1δ25 | 5′-GTCGACTCAGTAGGTGGTACGTCGTGG-3′ |
The PCR fragments were first cloned into the multiple cloning site of pcDNA-3.1/V5-His-TOPO (Invitrogen) and were then subcloned into the NcoI and XhoI sites of pGEX4T-3, generating the GST–CK1δ fusion proteins shown in Table 2.
Table 2. Generation of CK1δ-specific fragments, and nomenclature of pGEX–CK1δ plasmids and fusion proteins.
FP, fusion protein
| Primer pair | CK1δ amplified (bp) | pGEX vector name | GST–CK1δ fusion protein |
|---|---|---|---|
| CK1δ1+CK1δ2 | 154–912 | pGEX4T-3-CK1δ 52–304 | GST–CK1δ 52–304 (FP882) |
| CK1δ3+CK1δ4 | 445–1284 | pGEX4T-3-CK1δ 149–428 | GST–CK1δ 149–428 (FP894) |
| CK1δ5+CK1δ6 | 1–1125 | pGEX4T-3-CK1δ 1–375 | GST–CK1δ 1–375 (FP897) |
| CK1δ5+CK1δ7 | 1–1050 | pGEX4T-3-CK1δ 1–350 | GST–CK1δ 1–350 (FP898) |
| CK1δ17+CK1δ6 | 952–1125 | pGEX4T-3-CK1δ 318–375,S318A/T323A | GST–CK1δ 318–375,S318A/T323A (FP1001) |
| CK1δ18+CK1δ6 | 967–1125 | pGEX4T-3-CK1δ 323–375,T323A/T329A | GST–CK1δ 323–375,T323A/T329A (FP1003) |
| CK1δ19+CK1δ6 | 991–1125 | pGEX4T-3-CK1δ 331–375,S331A | GST–CK1δ 331–375,S331A (FP1004) |
| CK1δ8+CK1δ6 | 913–1125 | pGEX4T-3-CK1δ 305–375 | GST–CK1δ 305–375 (FP1006) |
| CK1δ9+CK1δ6 | 988–1125 | pGEX4T-3-CK1δ 330–375 | GST–CK1δ 330–375 (FP1011) |
| CK1δ8+CK1δ10 | 913–1071 | pGEX4T-3-CK1δ 305–357 | GST–CK1δ 305–357 (FP1012) |
| CK1δ11+CK1δ12 | 952–1053 | pGEX4T-3-CK1δ 318–351,S318A | GST–CK1δ 318–351,S318A (FP1013) |
| CK1δ8+CK1δ13 | 913–1029 | pGEX4T-3-CK1δ 305–343 | GST–CK1δ 305–343 (FP1014) |
| CK1δ14+CK1δ6 | 1021–1125 | pGEX4T-3-CK1δ 341–375 | GST–CK1δ 341–375 (FP1020) |
| CK1δ15+CK1δ16 | 1057–1125 | pGEX4T-3-CK1δ 353–375,S370A | GST–CK1δ 353–375,S370A (FP1021) |
| CK1δ15+CK1δ6 | 1057–1125 | pGEX4T-3-CK1δ 353–375 | GST–CK1δ 353–375 (FP1022) |
| CK1δ20+CK1δ21 | 1–1284 | pGEX2T-CK1δ 1–428,S370A | GST–CK1δ 1–428,S370A (FP1023) |
The plasmid pGEX2T–CK1δ 1–428,S370A, containing mutations at base pairs 1108 and 1109 (AG→GC) of rat CK1δ, was constructed using the QuikChange site-directed mutagenesis kit according to manufacturer's instructions (Stratagene). The plasmid pGEX2T–CK1δ 1–428 served as a template and the complementary primers used were CK1δ20 (5′ primer) and CK1δ21 (3′ primer) (see Tables 1 and 2).
Rat wild-type CK1δ was amplified by PCR using pGEX2T–CK1δ 1–428 (fusion protein 449) [34] and the primers CK1δ22 (5′ primer) and CK1δ23 (3′ primer) (see Tables 1 and 2), and was cloned into the pcDNA-3.1/V5-His-TOPO vector, generating pcDNA-3.1-HA (haemagglutinin)–CK1δ (fusion protein 1101).
Rat mutant CK1δ S370A was amplified by PCR using pGEX2T-CK1δ 1–428,S370A (fusion protein 1021) as a template and the primers CK1δ22 (5′ primer) and CK1δ23 (3′ primer) (see Tables 1 and 2), and was cloned into the pcDNA-3.1/V5-His-TOPO vector, generating pcDNA-3.1-HA–mutant CK1δ S370A (fusion protein 1102).
The kinase-dead rat CK1δ (mutant CK1δ K38M) was amplified by PCR using pGBKT7–CK1δ 1–428,K38M (fusion protein 737, [23]) as a template and the primers CK1δ24 (5′ primer/myc-tag) and CK1δ25 (3′ primer) (see Tables 1 and 2), and then was cloned into the pcDNA-3.1/V5-His-TOPO vector, generating pcDNA-3.1–myc–mutant CK1δ K38M.
All plasmids were sequenced at GATC Biotech.
Cell lines
Cells (MiaPaCa-2 [35], BxPc3 [36], Colo357 [37] and PancTu1 [38]) were grown in an 1:1 (v/v) mixture of DMEM (Dulbecco's modified Eagle's medium) and RPMI 1640 medium supplemented with 10% (v/v) FCS (fetal calf serum), 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified 5% CO2 atmosphere at 37 °C.
Antibodies
The rabbit polyclonal antibody against PKA (1 mg/ml; Upstate) and the mouse monoclonal CK1δ-specific antibody, 128A (4 mg/ml; ICOS Corporation), were used. HRP (horseradish peroxidase)-conjugated goat anti-(rabbit IgG) and goat anti-(mouse IgG) antibodies were purchased from GE Healthcare.
Cell treatment, labelling and cell lysis
MiaPaCa-2 cells were treated with 5 μM H89 (Calbiochem), a PKA-specific inhibitor, 20 μM forskolin (Calbiochem), a PKA activator, 10 μM St-Ht31 inhibitory peptide (Promega) or 10 μM St-Ht31P control peptide (Promega). Sub-confluent MiaPaCa-2 cells were labelled with 2 mCi of [32P]Pi/ml (GE Healthcare) in phosphate-free DMEM supplemented with 5% (v/v) phosphate-free FCS in the presence of either H89 (5 μM) or forskolin (20 μM) for 2 h. Cells were then washed in ice-cold PBS and lysed in sucrose lysis buffer [20 mM Tris/acetate, pH 7.0, 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X-100, 1 mM benzamidine, 4 μg/ml leupeptin, 30 μg/ml aprotinin and 0.1% (v/v) 2-mercaptoethanol] or NP40 (Nonidet P40) lysis buffer [50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 10% (v/v) glycerol, 1% NP4O, 5 mM DTT (dithiothreitol), 1 mM EDTA, 1 mM EGTA, 50 μM leupeptin and 30 μg/ml aprotinin].
Fractionation of cell extracts
MiaPaCa-2, BxPc3, Colo357 and PancTu1 cells were lysed in sucrose lysis buffer for 30 min at 4 °C. The cell lysate was centrifuged at 15000 g for 20 min at 4 °C, the supernatant was passed through a 0.40 μm-pore-size filter and 3 mg of total protein was applied to an anion-exchange column (Resource-Q) attached to an ETTAN purifier (GE Healthcare). The proteins were eluted with a linear ascending gradient between 0–1000 mM NaCl in 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 5% (v/v) glycerol, 0.04% Brij-35, 1 mM benzamidine, 4 μg/ml leupeptin and 0.1% (v/v) 2-mercaptoethanol.
Western blot analysis
Cells were lysed in NP40 lysis buffer, extracts were clarified by centrifugation at 15000 g for 20 min at 4 °C and the protein concentration of the lysates was determined using the BCA (bicinchoninic acid) protein assay (Pierce). From this determination, 75 μg of the protein extract was resolved on SDS/PAGE (12.5% gels) and transferred on to a Hybond C super nitrocellulose blotting membrane (GE Healthcare). The membranes were blocked in TBS containing 0.1% (v/v) Tween 20 and 5% (w/v) non-fat milk for 1 h before being probed with either the polyclonal rabbit antibody against PKA (1:1000 dilution) or with the monoclonal mouse antibody 128A (1:5000 dilution) against CK1δ. Immunocomplexes were detected by incubation for 45 min with HRP-conjugated goat anti-(rabbit IgG) or anti-(mouse IgG) (1:1000 dilution), followed by ECL® (enhanced chemiluminescence) detection (GE Healthcare).
Production and purification of GST fusion proteins
The production and purification of the GST fusion proteins (fusion proteins 267, 380, 449, 882, 894, 897, 898, 1001, 1003, 1004, 1006, 1011, 1012, 1013, 1014, 1020, 1021, 1022 and 1023) were carried out as described previously [23].
In vitro kinase assays and phosphopeptide analysis
In vitro kinase assays were carried out in the absence or presence of 5 μM H89 (PKA inhibitor), 200 nM deguelin (Akt inhibitor), or 1 μM calphostin C (PKCα inhibitor) as described previously [34]. Different GST–CK1δ fusion proteins, GST–p53 1–64 (fusion protein 267) and GST–p53 1–85,S4A/S6A/S9A (fusion protein 380), the CK1 peptide substrate (Asp-Asp-Asp-Glu-Glu-Ser-Ile-Thr-Arg-Arg) (Promega) [39] and β-casein (Sigma), were used as substrates. Recombinant PKA (Calbiochem), purified catalytic subunit of PKA from porcine heart (Sigma), recombinant human PKCα (amino acids 1–672) (Biomol), recombinant human Akt (amino acids 106–480) (Biomol), recombinant human CLK2 (Invitrogen) or single fractions of fractionated MiaPaCa-2 cell extracts were used as sources of enzyme. Phosphorylated proteins were resolved by SDS/PAGE and the protein bands were visualized by autoradiography. Where indicated, the phosphorylated protein bands were excised and quantified by Cherenkov counting.
Phosphopeptide analyses of in vitro and in vivo labelled proteins were performed as described earlier [23].
RNA synthesis
For microinjection experiments, run-off transcripts of rat wild-type HA-tagged CK1δ (plasmid 1101), HA-tagged mutant CK1δ S370A (plasmid 1102) and myc-tagged kinase-dead CK1δ K38M (plasmid 737) were synthesized using a T7 mMESSAGE mMACHINE® kit (Ambion).
Xenopus microinjection
Xenopus embryos were microinjected into both ventral blastomeres at the four-cell stage with the indicated amount of RNA. Secondary axis formation was analysed at stages 22 and 26 of development. Staging was determined as described by Nieuwkoop and Faber [40].
RESULTS
CK1δ activity differs in pancreatic tumour cell lines
It has been suggested previously [33] that the differences in the activity of CK1δ in extracts of various organs from young adult Balb/c mice might be influenced by site-specific phosphorylation of CK1δ. To investigate this further, the activity of CK1δ was analysed in fractionated extracts from apoptosis-resistant pancreatic tumour cell lines, all expressing similar protein levels of CK1δ. The kinase activity in fractionated BxPc3, MiaPaCa-2, PancTu1 and Colo357 cell extracts, which eluted at 220–240 mM NaCl and was inhibited by the CK1-specific inhibitor IC261 [41], differed in concentration by up to 6-fold (see Supplementary Figure 1S at http://www.BiochemJ.org/bj/406/bj4060389add.htm). These results suggested that post-translational modifications of CK1δ were involved in modulating the activity of CK1δ, and this was particularly true for site-specific phosphorylation.
Cellular kinases phosphorylate CK1δ at Ser370
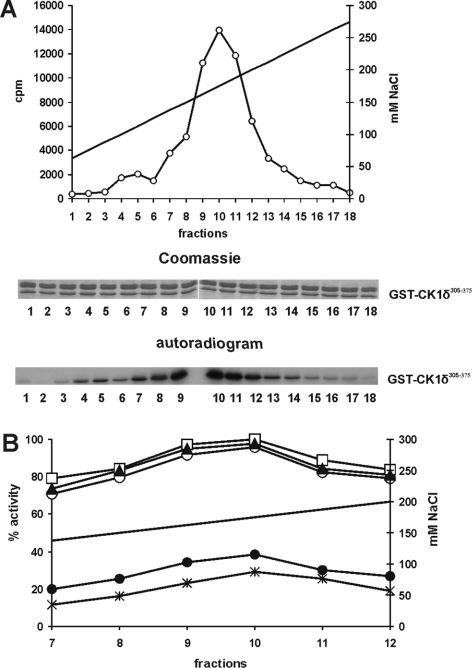
The C-terminal domain of CK1δ plays an important role in the regulation of the kinase activity of CK1δ. In order to identify cellular kinases which were able to phosphorylate CK1δ within its C-terminal domain, protein extracts of MiaPaCa-2 cells were prepared and fractionated by ion-exchange chromatography using a Resource-Q column. Each eluted fraction was tested for kinase activity using GST–CK1δ 305–375 as a substrate. A major kinase peak was detected in fractions eluted between 160 and 190 mM NaCl (Figure 1A), indicating that GST–CK1δ 305–375 can be phosphorylated by cellular kinases in vitro.
Figure 1. Characterization of CK1δCK activities in MiaPaCa-2 cells.
(A) Fractionation of CK1δCK activity present in MiaPaCa-2 cells by anion-exchange chromatography. A soluble protein extract (3 mg), derived from the lysis of MiaPaCa-2 cells, was fractionated on a Resouce-Q column using a linear ascending gradient of NaCl. Kinase assays were performed using equal amounts of GST–CK1δ 305–375 as a substrate and single fractions of fractionated MiaPaCa-2 cell extracts as the source of enzyme. Proteins were separated by SDS/PAGE and the phosphorylated proteins were detected by autoradiography. Quantification of phosphate incorporation into GST–p53 1–64 was measured by Cerenkov counting. A major kinase activity, which phosphorylated GST–CK1δ 305–375 (FP1006), eluted between 160 and 190 mM NaCl (fractions 9–11). (B) Localization of phosphorylation sites targeted by CK1δCK activiy. In vitro kinase assays were performed using fractions 7–12 of fractionated MiaPaCa-2 cell extracts as the source of enzyme activity and fusion proteins GST–CK1δ 305–375 (FP1006), GST–CK1δ 330–375 (FP1011), GST–CK1δ 305–357 (FP1012), GST–CK1δ 353–375 (FP1022) and GST–CK1δ 305–375,S370A (FP1021) as substrates. The highest cellular kinase activity (100%) was detected in fraction 10 when using GST–CK1δ 305–375 (FP1006) as substrate. □, GST–CK1δ 305–375. ○, CK1δ 330–375. ▲, CK1δ 353–375. *, CK1δ 353–375,S370A. ●, CK1δ 305–357. Solid line, NaCl concentration. FP, fusion protein.
In order to identify the CK1δCK (CK1δ C-terminal-targeted protein kinase)-dependent phosphorylation site or sites present within the C-terminus of CK1δ, additional GST–CK1δ fusion proteins consisting of amino acids 330–375, 305–357 and 353–375 were constructed (Tables 1 and 2) and used as substrates for phosphorylation by CK1δCK. Although cellular CK1δCK (phosphorylation activity for CK1δCK peaked in fraction 10) phosphorylated the truncated GST–CK1δ fusion proteins GST–CK1δ 330–375 and GST–CK1δ 353–375 to a similar extent (96 and 98% respectively) compared with GST–CK1δ 305–375 (100%), CK1δCK phosphorylated GST–CK1δ 305–357 approx. 38% compared with GST–CK1δ 305–375 (Figure 1B). Taken together, these results suggest that the major phosphorylation site or sites targeted by cellular kinases are localized within amino acids 353–375 of CK1δ.
A Scansite search (http://scansite.mit.edu) revealed that Ser 370 might be the major phosphorylation site within amino acids 353–375 of CK1δ, which is targeted by cellular kinases, including Akt, PKA, PKCα/β/γ/ζ and CLK2. Therefore GST–CK1δ 353–375,S370A, containing a serine to alanine mutation at position 370 was generated and used as a substrate for in vitro kinase assays. As shown in Figure 1(B), the degree of phosphorylation of GST–CK1δ 353–375,S370A was approx. 4-fold lower compared with that of GST–CK1δ 353–375. These results strongly suggest that the major phosphorylation site of CK1δ targeted by the CK1δCK activity is Ser370.
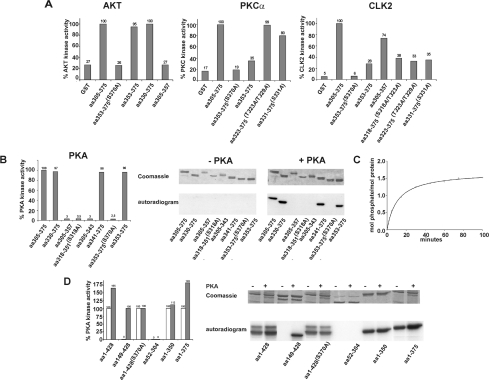
To prove the ability of PKA, Akt, PKCα and CLK2 to phosphorylate CK1δ at Ser370, several GST–CK1δ fusion proteins (GST–CK1δ 305–375, GST–CK1δ 353–375, GST–CK1δ 353–375,S370A, GST–CK1δ 305–357, GST–CK1δ 330–375, GST–CK1δ 318–375,S318A/T323A, GST–CK1δ 353–375,T323A/T329A and GST–CK1δ 331–375,S331A) were used in the in vitro kinase assays. The results shown in Figure 2(A) indicated that Akt predominantly phosphorylated Ser370 in vitro. In contrast, Ser370 was only weakly phosphorylated by PKCα and Clk2 in vitro. The major phosphorylation sites targeted by these two kinases were located within amino acids 305–353 of CK1δ (Figure 2A).
Figure 2. Phosphorylation of CK1δ by cellular kinases in vitro.
(A) Phosphorylation of CK1δ by Akt, PKCα and CLK2. In vitro kinase assays were performed using recombinant human PKCα (amino acids 1–672), recombinant human Akt (amino acids 106–480) and recombinant human CLK2 as sources of enzyme, and various GST–CK1δ fusion proteins (GST–CK1δ 305–375, GST–CK1δ 353–375, GST–CK1δ 353–375,S370A, GST–CK1δ 305–357, GST–CK1δ 330–375, GST–CK1δ 318–375,S318A/T323A, GST–CK1δ 353–375,T323A/T329A and GST–CK1δ 331–375,S331A) or GST as substrates. Quantification of phosphate incorporation of phosphorylated GST–CK1δ fusion proteins and GST was measured by Cerenkov counting. (B) Characterization of the site-specific phosphorylation of CK1δ by PKA. In vitro kinase assays were performed using the catalytic subunit of PKA purified from porcine heart as the source of enzyme and GST–CK1δ 305–375 (FP1006), GST–CK1δ 330–375 (FP1011), GST–CK1δ 305–357 (FP1012), GST–CK1δ 318–351,S318A (FP1013), GST–CK1δ 305–343 (FP1014), GST–CK1δ 341–375 (FP1020), GST–CK1δ 353–375,S370A (FP1021) and GST–CK1δ 353–375 (FP1022) as substrates. Equal amounts of GST–CK1δ fusion proteins were resolved by SDS/PAGE (12.5% gels) and stained with Coomassie Brilliant Blue. Quantification of phosphate incorporation of phosphorylated GST–CK1δ fusion proteins was measured by Cerenkov counting. (C) PKA phosphorylates one major residue in the C-terminal domain of CK1δ. GST–CK1δ 305–375 (FP1006) was phosphorylated in vitro by PKA for the indicated times. Up to 1.5 mol phosphate/mol protein was incorporated into the GST–CK1δ 305–375 fusion protein. (D) Phosphorylation of GST–CK1δ fusion proteins by PKA. In vitro kinase assays were performed using the catalytic subunit of PKA purified from porcine heart as the source of enzyme and GST–CK1δ 1–428 (FP449), GST–CK1δ 149–428 (FP894), GST–CK1δ 1–428,S370A (FP1023), GST–CK1δ 52–304 (FP882), GST–CK1δ1–350 (FP898) and GST–CK1δ 1–375 (FP897) as substrates. The autoradiogram, the Coomassie Blue-stained gel and the quantification analysis presented are of a representative experiment. Open bars, without PKA; closed bars, with PKA. aa, amino acid; FP, fusion protein.
Incubation of a series of GST–CK1δ fusion proteins (GST–CK1δ 305–375, GST–CK1δ 330–375, GST–CK1δ 305–357, GST–CK1δ 318–351,S318A, GST–CK1δ 305–343, GST–CK1δ 341–375, GST–CK1δ 353–375,S370A and GST–CK1δ 353–375) with the catalytic subunit of PKA purified from porcine heart and [γ-32P]ATP confirmed that Ser370 was the major phosphorylation site targeted by PKA in vitro (Figure 2B).
To prove the stoichiometry of the phosphorylation by PKA, in vitro kinase assays were performed using purified PKA as enzyme and GST–CK1δ 305–375 as substrate. The result of three independent experiments indicated that approx. 1.5 mol of phosphate were incorporated per mol of protein, suggesting that there is one major phosphorylation site within amino acids 305–375 of CK1δ that is phosphorylated by PKA in vitro (Figure 2C).
To examine whether additional serine and threonine residues of CK1δ are phosphorylated by PKA in vitro, the GST–CK1δ fusion proteins GST–CK1δ 1–428, GST–CK1δ 1–375, GST–CK1δ 1–350, GST–CK1δ 52–304 and GST–CK1δ 149–428 were used as substrates. All GST–CK1δ fusion proteins containing Ser370 (GST–CK1δ 1–428, GST–CK1δ 149–428 and GST–CK1δ 1–375) were phosphorylated by PKA. Even though GST–CK1δ 1–428 and GST–CK1δ 1–375 were highly autophosphorylated, the degree of phosphorylation increased up to 82% in the presence of purified PKA. In contrast with this, GST–CK1δ fusion proteins that did not encompass Ser370 (GST–CK1δ 1–350 and GST–CK1δ 52–304) were either weakly or not phosphorylated. Interestingly, when the GST–CK1δ 1–428,S370A fusion protein was used as a substrate, no change in the phosphorylation status of CK1δ was observed, underlining that PKA-mediated phosphorylation occurs predominantly at Ser370 in vitro (Figure 2D).
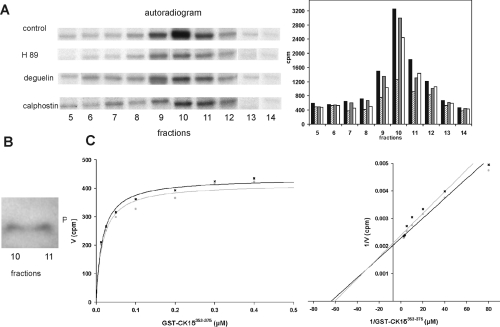
To gain more information regarding the identity of the CK1δCK activity, CK1δCK activity was assayed in the absence and presence of kinase inhibitors specific for PKA, PKCα or Akt, using GST–CK1δ 305–375 as the substrate. As demonstrated in Figure 3(A), a 65% reduction in CK1δCK activity was observed in the presence of the PKA-specific inhibitor H89 (5 μM), whereas, in the presence of the PKC-specific inhibitor calphostin C, (1 μM) or the Akt-specific inhibitor degeulin (200 nM), the reduction in CK1δCK activity was much lower (25 and 9% respectively).
Figure 3. PKA is the major CK1δCK activity present in the kinase peak fraction of fractionated MiaPaCa-2 cells.
(A) CK1δ-targeting kinase activities are mainly inhibited by the PKA-specific inhibitor H89. In vitro kinase assays were performed in the absence and presence of the PKA-specific inhibitor H89 (5 μM), the Akt-specific inhibitor degeulin (200 nM) or the PKC-specific inhibitor calphostin C (1 μM), using GST–CK1δ 305–375 (FP1006) as the substrate and fractions 5–14 of fractionated MiaPaCa-2 cell lysates as sources of kinase activity. Closed bars, control; hatched bars, H89; grey bars, degeulin; open bars, calphostin C. (B) PKA is present in the kinase peak fractions. PKA can be detected in the kinase peak fractions 10 and 11 of fractionated MiaPaCa-2 cell lysates by Western Blot analysis. (C) The kinetic properties of CK1δCK and PKA are nearly identical. To determine the kinetic parameters of CK1δCK and PKA, in vitro kinase assays were performed using GST–CK1δ 353–375 (FP1022) as a substrate. The results indicate that the Km and Vmax values of CK1δCK (grey dots) and PKA (black crosses) are nearly identical. FP, fusion protein.
As PKA seems to be the major CK1δCK-active kinase present in the kinase peak fractions (Figure 1A) and was clearly detected in the kinase peak (fractions 10 and 11) by Western blot analysis (Figure 3B), the kinetic parameters of CK1δCK activity were compared with those of recombinant PKA, using the GST–CK1δ 353–375 fusion protein as the substrate. The results indicate that the Km and Vmax values for CK1δCK and PKA are nearly identical (Figure 3C).
Taken together, all these experiments strongly point to PKA being the principal kinase responsible for the phosphorylation of CK1δ at Ser370.
Ser370 plays an important role in modulating the substrate phosphorylation of CK1δ in vitro
In order to examine the importance of Ser370 in the catalytic activity of CK1δ in vitro, the kinetic constants of phosphorylation of β-casein and GST–p53 1–64 by GST–CK1δ 1–428 and GST–CK1δ 1–428,S370A were calculated experimentally. GST–CK1δ 1–428,S370A had lower Km and higher Vmax values compared with those of GST–CK1δ 1–428 (see Supplementary Figure 2S at http://www.BiochemJ.org/bj/406/bj4060389add.htm). These observed differences revealed that CK1δ 1–428,S370A had a higher affinity for phosphorylating β-casein and GST–p53 1–64 compared with that of GST–CK1δ 1–428, which was also indicated by the efficiency parameter (Vmax/Km ratio) in Supplementary Figure 2S(C).
Phosphorylation of GST–CK1δ by PKA affects its activity in vitro
So far, our results clearly indicate that Ser370, which plays an important role in the modulation of the activity of CK1δ, is the major phosphorylation site targeted by PKA in vitro. To examine the influence of PKA phosphorylation on the activity of CK1δ, in vitro kinase assays were performed in the presence or absence of the catalytic subunit of PKA purified from porcine heart. GST–CK1δ 1–428 (fusion protein 449) was used as a source of enzyme activity. CK1δ was pre-incubated with [γ-32P]ATP for 15 min before the addition of GST–p53 1–64 (fusion protein 267) as a substrate, either alone or in combination with PKA for the indicated time points (see Supplementary Figure 3S, at http://www.BiochemJ.org/bj/406/bj4060389add.htm).
As shown in Supplementary Figure 3S, CK1δ-mediated phosphorylation of GST–p53 1–64 (fusion protein 267) was reduced up to 80% in the presence of PKA compared with that observed in the absence of the catalytic subunit of PKA. Phosphorylation of GST–p53 1–64 by PKA could be excluded as in vitro kinase assays demonstrated that PKA did not phosphorylate GST–p53 1–64 (results not shown).
Effects of CK1δ S370A in Xenopus embryogenesis
Different CK1 isoforms have been implicated in the Wnt signalling pathway. Considering the high affinity of CK1δ S370A for phosphorylating both β-casein and GST–p53 1–64 in vitro, we analysed the effects of this mutant on Wnt/β-catenin signalling in vivo using the Xenopus secondary axis induction assay.
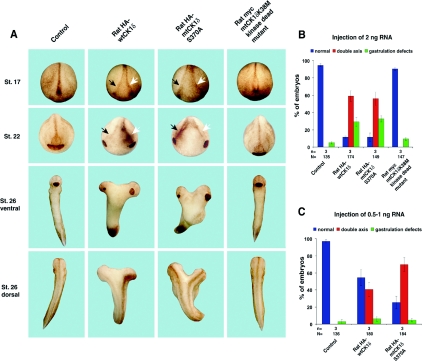
In vitro-transcribed RNA encoding HA-tagged rat wild-type CK1δ, mutant CK1δ S370A or myc-tagged mutant CK1δ K38M were microinjected into both ventral cells of four-cell-stage Xenopus embryos. The kinase-dead mutant CK1δ K38M did not perturb embryonic development, whereas high doses (2 ng) of either wild-type CK1δ or CK1δ S370A resulted in the formation of an ectopic dorsal axis (Figures 4A and 4B), as well as gastrulation movement defects. To identify any difference in the biological activity of both constructs, we lowered the concentration of injected RNA in subsequent experiments and selected for embryos which displayed a low gastrulation phenotype (not more than 10%, as this can occur non-specifically in egg batches of low quality or as an eventual result of injection of higher quantities of RNA). At lower amounts of microinjected mRNA, the secondary axis phenotype was demonstrated more in embryos expressing exogenous HA-tagged CK1δ S370A than in embryos expressing wild-type CK1δ (Figure 4C).
Figure 4. Secondary axis induction in X. laevis embryos.
X. laevis embryos were injected with wild-type or mutant CK1δ RNA at the four-cell stage as indicated. Evaluation of secondary axis formation was performed at stage (St.) 22 or 26. (A) Different views of embryos are shown. (B) Quantification of experiments showed that injection of higher amounts resulted in secondary axis formation and gastrulation movement defects in the case of wild-type CK1δ and CK1δ S370A. The kinase-dead mutant, CK1δ K38M, is inactive in this assay (n=3). N, total number of embryos used in all three experiments. (C) Injection of lower amounts of RNA and selection of embryo batches with low gastrulation movement phenotype indicated that CK1δ S370A has a higher capacity to induce secondary axis formation compared with wild-type CK1δ (n=3). N, total number of embryos used in all three experiments.
PKA phosphorylates CK1δ in vivo
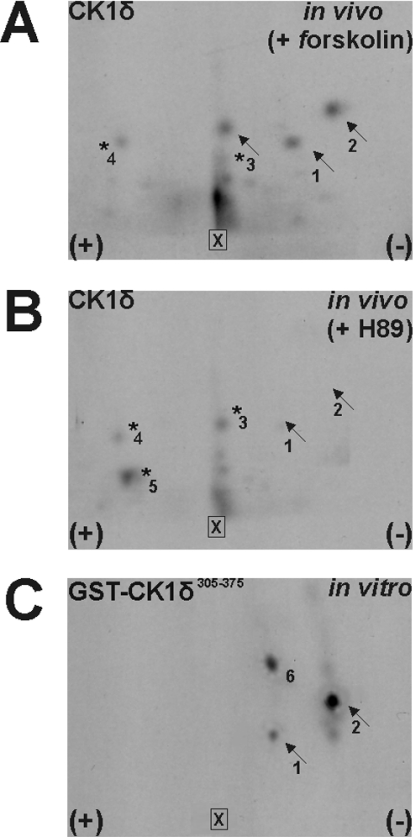
To examine whether CK1δ is phosphorylated by PKA in tissue culture cells, experiments were performed on the basis that modulation of the activity of PKA should influence the phosphorylation status of CK1δ. For this purpose, MiaPaCa-2 cells were treated either with 5 μM H89 (PKA-specific inhibitor) or with 20 μM forskolin (PKA activator) in the presence of 2 mCi [32P]Pi for 2 h. Cells were lysed and phosphorylated CK1δ was immunoprecipitated, isolated, digested with trypsin and oxidized. The resulting tryptic phosphopeptides were separated by two-dimensional tryptic phosphopeptide mapping and compared with the phosphopeptides derived from the GST–CK1δ 305–375 fusion protein, which was phosphorylated by PKA in vitro.
Phosphopeptides 1 and 2 were present in CK1δ isolated from MiaPaCa-2 cells treated with forskolin (Figure 5A) and in GST–CK1δ 305–375 (Figure 5C), whereas these peptides were not detectable in CK1δ isolated from MiaPaCa-2 cells that had been treated with H89 (Figure 5B), suggesting that PKA is the cellular kinase responsible for the phosphorylation of amino acid residues within these two peptides. Other migrated peptides (labelled peptides 3 and 4) could be detected in the phosphopeptide analyses of CK1δ isolated from both forskolin- and H89-treated cells, indicating the existence of additional phosphorylation sites.
Figure 5. Phosphopeptide analyses of CK1δ and GST–CK1δ 305–375.
GST–CK1δ 305–375 and [32P]-labelled CK1δ isolated from MiaPaCa-2 cells were treated with either 20 μM forskolin (A) or 5 μM H89 (B) for 2 h and were oxidized and digested with trypsin (see Materials and methods section). In each case, the tryptic phosphopeptides were analysed by two-dimensional phosphopeptide mapping. The phosphopeptides are numbered from 1 to 6. Arrows indicate those peptides that are identical in the in vitro and the in vivo maps. (A) CK1δ-specific phosphopeptides from forskolin-treated MiaPaCa-2 cells. (B) Phosphopeptides of CK1δ from H89-treated MiaPaCa-2 cells. (C) Phosphopeptides from the GST–CK1δ 305–375 fusion protein phosphorylated by PKA in vitro. ×, loading point.
Subcellular localization of PKA is important for modulating the activity of CK1δ
Compartmentalization of signal transduction molecules, such as kinases and phosphatases, occurs by anchoring of proteins to scaffold proteins. This allows substrate phosphorylation and dephosphorylation in conjunction with various kinases and phosphatases, which results in compartment-specific regulation of the substrate.
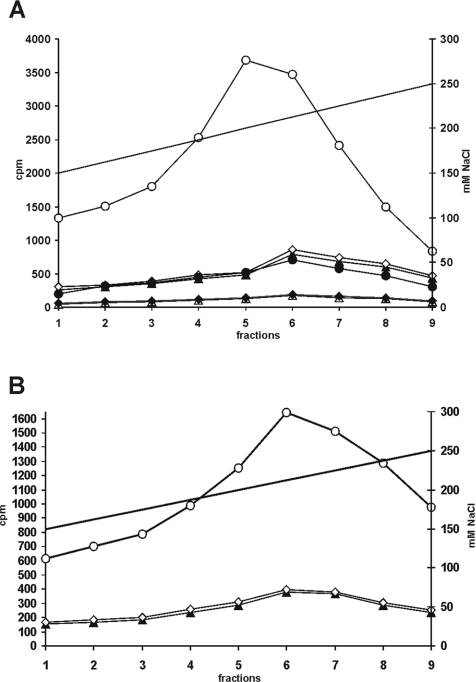
It has been reported that the interaction of PKA and CK1δ with AKAP450 docks both kinases to centrosomes [22,42,43], along with other signal transduction molecules. The binding of PKA to AKAPs can be blocked by the membrane-permeant AKAP-inhibitory peptide St-Ht31 [42,44]. Therefore we investigated whether disruption of the PKA–AKAP complex by the anchoring-disrupting peptide St-Ht31 [45,46] could influence the activity levels of CK1δ. For this purpose, MiaPaCa-2 cells were treated with 10 μM of either St-Ht31 or St-Ht31P control peptide [47] for 3 h. Cell lysates were fractionated by anion-exchange chromatography and the CK1δ kinase activity was determined. When the CK1 substrate peptide (Figure 6B) or GST–p53 1–64 (fusion protein 267) (Figure 6A) were used as substrates, the CK1δ activity increased 3–6-fold in cells treated with the St-Ht31 inhibitory peptide compared with untreated cells and cells treated with the St-Ht31P control peptide (Figure 6).
Figure 6. Inhibition of the interaction of PKA with AKAPs is accompanied by an increase in CK1δ activity.
Equal amounts of protein (3 mg) from MiaPaCa-2 cell lysates which had been untreated, treated with 10 μM St-Ht31 control peptide or with 10 μM St-Ht31-specific inhibitory peptide were fractionated (see Materials and methods section). The proteins were eluted using a linear gradient of increasing NaCl concentrations (represented as a solid line). (A) The CK1δ-specific kinase activity was determined using GST–p53 1–64 (FP267) and GST–p53 1–84,S4A/S6A/S9A fusion proteins (FP380) as substrates. When GST–p53 1–64 was used as a substrate: ○, cells treated with St-Ht31 inhibitory peptide; ◇, cells treated with St-Ht31 control; ▲, untreated cells. When GST–p53 1–84,S4A/S6A/S9A was used as a substrate: ●, cells treated with St-Ht31 inhibitory peptide; ♦, cells treated with St-Ht31 control; △, untreated cells. (B) CK1δ-specific kinase activity was also determined using the CK1 peptide substrate Asp-Asp-Asp-Glu-Glu-Ser-Ile-Thr-Arg-Arg. ○, Cells treated with St-Ht31 inhibitory peptide; ◇, cells treated with St-Ht31 control peptide; ▲, untreated cells. FP, fusion protein.
Differences in the CK1δ kinase activity were observed in cells treated with the St-Ht31 inhibitory peptide when GST–p53 1–85,S4A/S6A/S9A (fusion protein 380) was used as a substrate, compared with untreated cells and cells treated with the St-Ht31P control peptide (Figure 6A). However, as expected, the CK1δ-mediated phosphorylation of GST–p53 1–85,S4A/S6A/S9A was significantly lower compared with that of GST–p53 1–64, owing to the mutation of the majority of the CK1δ phosphorylation sites (S4A/S6A/S9A) within the N-terminal domain of p53.
DISCUSSION
CK1δ, a member of the CK1 family, is involved in the regulation of many different cellular processes, including cell proliferation and cell death. Therefore several mechanisms ensure a tight regulation of CK1δ at different levels [15,24,26,28,48,49].
Using a biochemical approach to clarify the role of cellular kinases in modulating CK1δCK activity, MiaPaCa-2 cell lysates were fractionated and the kinase activity was detected in fractions eluted in 160–190 mM NaCl. The use of various C-terminal-truncated wild-type and mutant GST–CK1δ fusion proteins as substrates revealed that Ser370 is the main residue targeted by CK1δCK for phosphorylation. Ser370 is a potential phospho-acceptor site for phosphorylation by several putative kinases, including Akt, PKA, CLK2 and PKCα. In vitro kinase assays using various GST–CK1δ fusion proteins as substrates revealed that PKA and Akt mainly phosphorylate Ser370, whereas the key targeted sites of PKCα and CLK2 lie within amino acids 305–353 of CK1δ. However, additional experiments are necessary to identify these phosphorylation sites targeted by PKCα and CLK2 and to elucidate their role in regulating the activity of CK1δ.
Focussing on the phosphorylation of Ser370, our results strongly suggest that this serine residue is predominantly phosphorylated by PKA. PKA co-eluted with the peak of the CK1δCK activity, as verified by Western blot analysis, suggesting that PKA provides CK activity. The PKA-specific inhibitor H89 inhibited the kinase activity present in the kinase peak fraction up to 65%, whereas in the presence of the Akt-specific inhibitor degeulin or in the presence of the PKC-specific inhibitor calphostin C a much weaker reduction was observed in kinase activity. Comparison of the kinetic parameters of recombinant PKA with those of CK1δCK present in the kinase peak fraction indicated nearly identical parameters. The purified catalytic subunit of PKA, as well as recombinant PKA, phosphorylated all GST–CK1δ fusion proteins containing Ser370 in vitro, whereas all GST–CK1δ fusion proteins lacking Ser370, or containing a serine to alanine substitution at Ser370, were not phosphorylated, suggesting that PKA is responsible for the phosphorylation of Ser370. Furthermore, time-course experiments revealed that there is one major phosphorylation site for PKA within amino acids 305–375. The presence of arginine and lysine residues at positions n−3 and n−2 on the N-terminal side of Ser370 (R265ERKVS370) resembles a perfect consensus recognition motif for PKA [50], thereby favouring Ser370 as the main phosphorylation site targeted by PKA. Moreover, comparison of our phosphopeptide analyses suggested that CK1δ is phosphorylated in vivo at the same sites which are phosphorylated by PKA in vitro.
In addition, phosphorylation of CK1δ by PKA reduced the ability of CK1δ to phosphorylate GST–p53 1–64 up to 80%. This result indicated that site-specific phosphorylation may be involved in the modulation of CK1δ activity.
The importance of Ser370 for the substrate phosphorylation of CK1δ was indicated by comparing the kinetic parameters of GST–CK1δ 1–428 with those of GST–CK1δ 1–428,S370A. Our results show that the GST–CK1δ 1–428,S370A mutant exhibits lower Km and higher Vmax values when compared with those of GST–CK1δ 1–428, when β-casein and GST–p53 1–64 were used as substrates.
Furthermore, the role of Ser370 appears to be physiologically important, since it has been shown that mutation of this site can interfere with the normal development of Xenopus embryos. Expression of the mutant kinase (CK1δ S370A) in Xenopus embryos resulted in a higher rate of ectopic dorsal axis formation compared with the expression of wild-type CK1δ, whereas the kinase-dead mutant (CK1δ K38M) failed to induce axis formation.
In addition, our results strongly support the idea that CK1δ and PKA interact physiologically. This is due to the observation that inhibition of the interaction of PKA with AKAPs by the St-Ht31 inhibitory peptide [45,51] resulted in an increase in CK1δ activity. The increased activity of CK1δ could be explained by changes in the phosphorylation status of CK1δ. Loss of the interaction between CK1δ and PKA could lead to reduced phosphorylation of Ser370. Furthermore, cellular phosphatases associated with AKAPs [52,53] could more efficiently dephosphorylate the autophosphorylation sites of CK1δ in the absence of PKA, thereby activating CK1δ. However, St-Ht31 treatment of cells can also decrease the activity of signalling molecules, as it has been shown that St-Ht31 could disrupt the activity of RhoA [54].
In summary, these results show for the first time that CK1δ is phosphorylated by several kinases, and that site-specific phosphorylation, especially at Ser370 by PKA, plays an important role in modulating the activity of CK1δ.
Online data
Acknowledgments
This work was supported by the Deutsche Krebshilfe, Dr Mildred Scheel Stiftung (10-2237-KN3) to U.K. Work in the laboratory of M. K. is supported by funding (SFB 497/A6) from the DFG (Deutsche Forschungsgemeinschaft). We thank Tony DeMaggio (ICOS Corporation) for providing us with the CK1δ-specific monoclonal antibody 128A, David Meek (Biomedical Research Centre, Ninewells Hospital and Medical School, University of Dundee, Dundee, Scotland, U.K.) for pGEX2T–CK1δ (rat), pGEX–p53 1–64 (mouse) and pGEX–p53 1–84,S4A/S6A/S9A (mouse), Tilo Patschinsky for helpful discussions and reading the manuscript prior to submission, and Annette Blatz and Corinna Patzina for technical assistance.
References
- 1.Knippschild U., Gocht A., Wolff S., Huber N., Lohler J., Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signalling. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Flotow H., Graves P. R., Wang A. Q., Fiol C. J., Roeske R. W., Roach P. J. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 3.Flotow H., Roach P. J. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 1991;266:3724–3727. [PubMed] [Google Scholar]
- 4.Meggio F., Perich J. W., Reynolds E. C., Pinna L. A. A synthetic β-casein phosphopeptide and analogues as model substrates for casein kinase-1, a ubiquitous, phosphate directed protein kinase. FEBS Lett. 1991;283:303–306. doi: 10.1016/0014-5793(91)80614-9. [DOI] [PubMed] [Google Scholar]
- 5.Pulgar V., Marin O., Meggio F., Allende C. C., Allende J. E., Pinna L. A. Optimal sequences for non-phosphate-directed phosphorylation by protein kinase CK1 (casein kinase-1): a re-evaluation. Eur. J. Biochem. 1999;260:520–526. doi: 10.1046/j.1432-1327.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- 6.Marin O., Bustos V. H., Cesaro L., Meggio F., Pagano M. A., Antonelli M., Allende C. C., Pinna L. A., Allende J. E. A noncanonical sequence phosphorylated by casein kinase 1 in β-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustos V. H., Marin O., Meggio F., Cesaro L., Allende C. C., Allende J. E., Pinna L. A. Generation of protein kinase Ck1α mutants which discriminate between canonical and non-canonical substrates. Biochem. J. 2005;391:417–424. doi: 10.1042/BJ20050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross S. D., Simerly C., Schatten G., Anderson R. A. A casein kinase I isoform is required for proper cell cycle progression in the fertilized mouse oocyte. J. Cell Sci. 1997;110:3083–3090. doi: 10.1242/jcs.110.24.3083. [DOI] [PubMed] [Google Scholar]
- 9.Hoekstra M. F., DeMaggio A. J., Dhillon N. Genetically identified protein kinases in yeast. II: DNA metabolism and meiosis. Trends Genet. 1991;7:293–297. doi: 10.1016/0168-9525(91)90311-D. [DOI] [PubMed] [Google Scholar]
- 10.Santos J. A., Logarinho E., Tapia C., Allende C. C., Allende J. E., Sunkel C. E. The casein kinase 1α gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J. Cell Sci. 1996;109:1847–1856. doi: 10.1242/jcs.109.7.1847. [DOI] [PubMed] [Google Scholar]
- 11.Sakaguchi K., Saito S., Higashimoto Y., Roy S., Anderson C. W., Appella E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian D., Griffith J. D. Modulation of p53 binding to Holliday junctions and 3-cytosine bulges by phosphorylation events. Biochemistry. 2005;44:2536–2544. doi: 10.1021/bi048700u. [DOI] [PubMed] [Google Scholar]
- 13.Brockman J. L., Gross S. D., Sussman M. R., Anderson R. A. Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9454–9458. doi: 10.1073/pnas.89.20.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behrend L., Stoter M., Kurth M., Rutter G., Heukeshoven J., Deppert W., Knippschild U. Interaction of casein kinase 1δ (CK1δ) with post-Golgi structures, microtubules and the spindle apparatus. Eur. J. Cell Biol. 2000;79:240–251. doi: 10.1078/s0171-9335(04)70027-8. [DOI] [PubMed] [Google Scholar]
- 15.Milne D. M., Looby P., Meek D. W. Catalytic activity of protein kinase CK1δ (casein kinase 1δ) is essential for its normal subcellular localization. Exp. Cell Res. 2001;263:43–54. doi: 10.1006/excr.2000.5100. [DOI] [PubMed] [Google Scholar]
- 16.Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., Schwickart M., Mechtler K., Shirahige K., Zachariae W., Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Beyaert R., Vanhaesebroeck B., Declercq W., Van Lint J., Vandenabele P., Agostinis P., Vandenheede J. R., Fiers W. Casein kinase-1 phosphorylates the p75 tumor necrosis factor receptor and negatively regulates tumor necrosis factor signaling for apoptosis. J. Biol. Chem. 1995;270:23293–23299. doi: 10.1074/jbc.270.40.23293. [DOI] [PubMed] [Google Scholar]
- 18.Desagher S., Osen-Sand A., Montessuit S., Magnenat E., Vilbois F., Hochmann A., Journot L., Antonsson B., Martinou J. C. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y., Qin S., Atangan L. I., Molina Y., Okawa Y., Arpawong H. T., Ghosn C., Xiao J. H., Vuligonda V., Brown G., Chandraratna R. A. Casein kinase 1α interacts with retinoid X receptor and interferes with agonist-induced apoptosis. J. Biol. Chem. 2004;279:30844–30849. doi: 10.1074/jbc.M404651200. [DOI] [PubMed] [Google Scholar]
- 20.Izeradjene K., Douglas L., Delaney A. B., Houghton J. A. Casein kinase I attenuates tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by regulating the recruitment of fas-associated death domain and procaspase-8 to the death-inducing signaling complex. Cancer Res. 2004;64:8036–8044. doi: 10.1158/0008-5472.CAN-04-0762. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie J. A., Riento K., Ridley A. J. Casein kinase Iϵ associates with and phosphorylates the tight junction protein occludin. FEBS Lett. 2006;580:2388–2394. doi: 10.1016/j.febslet.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 22.Sillibourne J. E., Milne D. M., Takahashi M., Ono Y., Meek D. W. Centrosomal anchoring of the protein kinase CK1δ mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J. Mol. Biol. 2002;322:785–797. doi: 10.1016/s0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- 23.Wolff S., Xiao Z., Wittau M., Sussner N., Stoter M., Knippschild U. Interaction of casein kinase 1δ (CK1δ) with the light chain LC2 of microtubule associated protein 1A (MAP1A) Biochim. Biophys. Acta. 2005;1745:196–206. doi: 10.1016/j.bbamcr.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Yin H., Laguna K. A., Li G., Kuret J. Dysbindin structural homologue CK1BP is an isoform-selective binding partner of human casein kinase-1. Biochemistry. 2006;45:5297–5308. doi: 10.1021/bi052354e. [DOI] [PubMed] [Google Scholar]
- 25.Carmel G., Leichus B., Cheng X., Patterson S. D., Mirza U., Chait B. T., Kuret J. Expression, purification, crystallization, and preliminary x-ray analysis of casein kinase-1 from Schizosaccharomyces pombe. J. Biol. Chem. 1994;269:7304–7309. [PubMed] [Google Scholar]
- 26.Graves P. R., Roach P. J. Role of COOH-terminal phosphorylation in the regulation of casein kinase I δ. J. Biol. Chem. 1995;270:21689–21694. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- 27.Gietzen K. F., Virshup D. M. Identification of inhibitory autophosphorylation sites in casein kinase Iϵ. J. Biol. Chem. 1999;274:32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 28.Cegielska A., Gietzen K. F., Rivers A., Virshup D. M. Autoinhibition of casein kinase Iϵ (CKIϵ) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 1998;273:1357–1364. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- 29.Swiatek W., Tsai I. C., Klimowski L., Pepler A., Barnette J., Yost H. J., Virshup D. M. Regulation of casein kinase Iϵ activity by Wnt signaling. J. Biol. Chem. 2004;279:13011–13017. doi: 10.1074/jbc.M304682200. [DOI] [PubMed] [Google Scholar]
- 30.Chergui K., Svenningsson P., Greengard P. Physiological role for casein kinase 1 in glutamatergic synaptic transmission. J. Neurosci. 2005;25:6601–6609. doi: 10.1523/JNEUROSCI.1082-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu F., Ma X. H., Ule J., Bibb J. A., Nishi A., DeMaggio A. J., Yan Z., Nairn A. C., Greengard P. Regulation of cyclin-dependent kinase 5 and casein kinase 1 by metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11062–11068. doi: 10.1073/pnas.191353898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maritzen T., Lohler J., Deppert W., Knippschild U. Casein kinase Iδ (CKIδ) is involved in lymphocyte physiology. Eur. J. Cell Biol. 2003;82:369–378. doi: 10.1078/0171-9335-00323. [DOI] [PubMed] [Google Scholar]
- 34.Knippschild U., Oren M., Deppert W. Abrogation of wild-type p53 mediated growth-inhibition by nuclear exclusion. Oncogene. 1996;12:1755–1765. [PubMed] [Google Scholar]
- 35.Yunis A. A., Arimura G. K., Russin D. J. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. Int. J. Cancer. 1977;19:218–235. doi: 10.1002/ijc.2910190118. [DOI] [PubMed] [Google Scholar]
- 36.Tan M. H., Nowak N. J., Loor R., Ochi H., Sandberg A. A., Lopez C., Pickren J. W., Berjian R., Douglass H. O., Jr, Chu T. M. Characterization of a new primary human pancreatic tumor line. Cancer Invest. 1986;4:15–23. doi: 10.3109/07357908609039823. [DOI] [PubMed] [Google Scholar]
- 37.Morgan R. T., Woods L. K., Moore G. E., Quinn L. A., McGavran L., Gordon S. G. Human cell line (COLO 357) of metastatic pancreatic adenocarcinoma. Int. J. Cancer. 1980;25:591–598. doi: 10.1002/ijc.2910250507. [DOI] [PubMed] [Google Scholar]
- 38.Moore P. S., Sipos B., Orlandini S., Sorio C., Real F. X., Lemoine N. R., Gress T., Bassi C., Kloppel G., Kalthoff H., et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 39.Agostinis P., Pinna L. A., Meggio F., Marin O., Goris J., Vandenheede J. R., Merlevede W. A synthetic peptide substrate specific for casein kinase I. FEBS Lett. 1989;259:75–78. doi: 10.1016/0014-5793(89)81498-x. [DOI] [PubMed] [Google Scholar]
- 40.Nieuwkoop P. D., Faber J. Normal Table of Xenopus laevis (Daudin): a Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis Hubrecht Laboratory. Utrecht. 2003 [Google Scholar]
- 41.Mashhoon N., DeMaggio A. J., Tereshko V., Bergmeier S. C., Egli M., Hoekstra M. F., Kuret J. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J. Biol. Chem. 2000;275:20052–20060. doi: 10.1074/jbc.M001713200. [DOI] [PubMed] [Google Scholar]
- 42.Carr D. W., Stofko-Hahn R. E., Fraser I. D., Cone R. D., Scott J. D. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J. Biol. Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- 43.Glantz S. B., Amat J. A., Rubin C. S. cAMP signaling in neurons: patterns of neuronal expression and intracellular localization for a novel protein, AKAP 150, that anchors the regulatory subunit of cAMP-dependent protein kinase IIβ. Mol. Biol. Cell. 1992;3:1215–1228. doi: 10.1091/mbc.3.11.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hausken Z. E., Coghlan V. M., Hastings C. A., Reimann E. M., Scott J. D. Type II regulatory subunit (RII) of the cAMP-dependent protein kinase interaction with A-kinase anchor proteins requires isoleucines 3 and 5. J. Biol. Chem. 1994;269:24245–24251. [PubMed] [Google Scholar]
- 45.Carr D. W., Hausken Z. E., Fraser I. D., Stofko-Hahn R. E., Scott J. D. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J. Biol. Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 46.Stokka A. J., Gesellchen F., Carlson C. R., Scott J. D., Herberg F. W., Tasken K. Characterization of A-kinase anchoring disruptors using a solution based assay. Biochem. J. 2006;400:493–499. doi: 10.1042/BJ20060962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijayaraghavan S., Goueli S. A., Davey M. P., Carr D. W. Protein kinase A-anchoring inhibitor peptides arrest mammalian sperm motility. J. Biol. Chem. 1997;272:4747–4752. doi: 10.1074/jbc.272.8.4747. [DOI] [PubMed] [Google Scholar]
- 48.Fish K. J., Cegielska A., Getman M. E., Landes G. M., Virshup D. M. Isolation and characterization of human casein kinase Iϵ (CKI), a novel member of the CKI gene family. J. Biol. Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- 49.Longenecker K. L., Roach P. J., Hurley T. D. Crystallographic studies of casein kinase Iδ toward a structural understanding of auto-inhibition. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:473–475. doi: 10.1107/s0907444997011724. [DOI] [PubMed] [Google Scholar]
- 50.Songyang Z., Blechner S., Hoagland N., Hoekstra M. F., Piwnica-Worms H., Cantley L. C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 51.Ruehr M. L., Zakhary D. R., Damron D. S., Bond M. Cyclic AMP-dependent protein kinase binding to A-kinase anchoring proteins in living cells by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. J. Biol. Chem. 1999;274:33092–33096. doi: 10.1074/jbc.274.46.33092. [DOI] [PubMed] [Google Scholar]
- 52.Bridges D., Macdonald J. A., Wadzinski B., Moorhead G. B. Identification and characterization of D-AKAP1 as a major adipocyte PKA and PP1 binding protein. Biochem. Biophys. Res. Commun. 2006;346:351–357. doi: 10.1016/j.bbrc.2006.05.138. [DOI] [PubMed] [Google Scholar]
- 53.Kuntziger T., Rogne M., Folstad R. L., Collas P. Association of PP1 with its regulatory subunit AKAP149 is regulated by serine phosphorylation flanking the RVXF motif of AKAP149. Biochemistry. 2006;45:5868–5877. doi: 10.1021/bi060066s. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Chen Y., Chen M., Xu W. AKAPs competing peptide HT31 disrupts the inhibitory effect of PKA on RhoA activity. Oncol. Rep. 2006;16:755–761. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.