Abstract
A proteome analysis of Arabidopsis thaliana thylakoid-associated polysome nascent chain complexes was performed to find novel proteins involved in the biogenesis, maintenance and turnover of thylakoid protein complexes, in particular the PSII (photosystem II) complex, which exhibits a high turnover rate. Four unknown proteins were identified, of which TLP18.3 (thylakoid lumen protein of 18.3 kDa) was selected for further analysis. The Arabidopsis mutants (SALK_109618 and GABI-Kat 459D12) lacking the TLP18.3 protein showed higher susceptibility of PSII to photoinhibition. The increased susceptibility of ΔTLP18.3 plants to high light probably originates from an inefficient reassembly of PSII monomers into dimers in the grana stacks, as well as from an impaired turnover of the D1 protein in stroma exposed thylakoids. Such dual function of the TLP18.3 protein is in accordance with its even distribution between the grana and stroma thylakoids. Notably, the lack of the TLP18.3 protein does not lead to a severe collapse of the PSII complexes, suggesting a redundancy of proteins assisting these particular repair steps to assure functional PSII. The ΔTLP18.3 plants showed no clear visual phenotype under standard growth conditions, but when challenged by fluctuating light during growth, the retarded growth of ΔTLP18.3 plants was evident.
Keywords: Arabidopsis thaliana, D1 protein, photosystem II (PSII), polysome nascent chain complex, proteome analysis, thylakoid lumen
Abbreviations: BN, blue native; Chl, chlorophyll; cyt, cytochrome; 2D, two-dimensional; DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; DMBQ, 2,6-dimethyl-p-benzoquinone; DUF, domain of unknown function; FNR, ferredoxin–NADP+ reductase; GL, growth light; HL, high light; IEF, isoelectric focusing; LHC, light-harvesting complex; MALDI–TOF, matrix-assisted laser-desorption-ionization time-of-flight; OEC, oxygen evolving complex; PAM, pulse amplitude modulation; PSI, photosystem I; PSII, photosystem II; T-DNA, transfer DNA; TMH, transmembrane helix; TLP18.3, thylakoid lumen protein of 18.3 kDa; TLP18.3*, truncated form of TLP18.3; WT, wild-type
INTRODUCTION
Thylakoid membrane-embedded multi-subunit protein complexes PSI (photosystem I), PSII (photosystem II), cyt (cytochrome) b6f and ATP synthase are responsible for light harvesting and solar energy transduction into chemical energy. Although the photosynthetic apparatus and light-driven electron transport have been studied extensively, the biosynthesis and maintenance of these complexes have remained largely unknown. PSII performs water oxidation and, as the most oxidizing protein complex in nature, it exerts photodamaging effects on its own protein components in a process known as PSII photoinhibition [1–4]. The light-induced photoinhibition of PSII results in an irreversible damage of the D1 protein, one of the heterodimeric polypeptides of the PSII core, and as a consequence the PSII core is subjected to a multi-step repair cycle, thereby maintaining the function of the photosynthetic light reactions [3,5].
Only a few proteins related to the regulatory network of the PSII complex have been characterized so far. However, the accomplishment of the Arabidopsis genome sequencing project and multiple proteomic studies localizing unknown proteins to different chloroplast compartments [6–9] have established a basis for identification of novel proteins possibly associated with the dynamics of the PSII complex. One such protein, PsbS, is involved in energy-dependent quenching of excess light energy, thus partially protecting the PSII complex against severe photoinhibition under fluctuating HL (high light) conditions [10]. Despite the mechanisms protecting the D1 protein from excess light, the light-induced damage and repair do constantly take place. These processes include: (i) a reversible phosphorylation of the PSII core proteins at least partially catalysed by the Stn8 protein kinase [11], although the phosphatases involved in the process still remain unknown; (ii) migration of the damaged PSII core from grana stacks to stroma-exposed membranes; (iii) degradation of the D1 protein by the FtsH and DegP proteases [12,13]; (iv) de novo synthesis of D1 protein and its co-translational insertion into PSII core in stroma-exposed membranes [14], followed by; (v) the re-assembly of functional PSII. Several proteins have been recognized that regulate the stability and translation of the psbA mRNA encoding the D1 protein, as well as the D1 processing protease CtpA [15–18]. On the contrary, only a few auxiliary proteins, such as Psb29, Lpa1, HCF136 and Psb27, have been identified to be essential in the PSII assembly process [19–22].
To gain further insight into the biogenesis, maintenance and turnover of the thylakoid protein complexes, we have taken a proteomic approach focusing on thylakoid-associated polysome nascent chain complexes, referred hereafter as polysomes. Translation of most proteins encoded by the chloroplast genome takes place on ribosomes attached to the stroma-exposed membranes [23]. Of all thylakoid proteins the PSII core protein D1 has the highest turnover rate [24], and therefore the proteome analysis of polysomes is a promising approach in attempts to find novel proteins particularly involved in the PSII repair cycle. In the present study, 40 proteins of Arabidopsis polysomes were determined, of which five possibly function as auxiliary proteins in the PSII repair. Of those proteins At1g54780 was enriched in the polysomes and was selected for further analysis. The At1g54780 protein, earlier unambiguously demonstrated to be located on the lumenal side of the thylakoid membrane [8,9], was named as TLP18.3 (thylakoid lumen protein of 18.3 kDa), and two Arabidopsis TLP18.3 T-DNA (transfer DNA) insertion mutant lines were subjected to detailed characterization.
EXPERIMENTAL
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia WT (wild-type) plants and homozygous TLP18.3 T-DNA insertion mutants on Columbia background (SALK_109618 and GABI-Kat 459D12) were used in the experiments [25,26]. Information about insertion mutants was obtained from the Salk Institute Genomic Analysis Laboratory website (http://signal.salk.edu) and from the GABI-Kat Arabidopsis thaliana T-DNA mutagenized population website (http://www.gabi-kat.de). Plants were analysed by PCR using primers specific to the flanking sequences either on the left (LP) or on the right (RP) side of At1g54780 gene together with a primer within the T-DNA insert (LB). The following primers were used (5′→3′): LP1 (GTCCGTTGCTAGTACTGCACC), RP1 (CTCCGGTCCTAAACACCTCTC) and LB1 (TGGTTCACGTAGTGGGCCATCG) for analysis of SALK_109618 plants, and LP2 (GGAAGGAGCTATTACTGGTGG), RP2 (CGAAAAACCGTTAATGGCGTG) and LB2 (CCATATTGACCATCATACTCATTGC) for analysis of GABI-Kat 459D12 plants. Immunoblot analysis using a peptide antibody raised against the TLP18.3 protein was carried out to confirm that the TLP18.3 protein was lacking from the selected mutant plants.
Plants were grown in GL (growth light) in a growth chamber under a photon flux density of 150 μmol of photons·m−2·s−1 in 8 h light/16 h dark regime at 23 °C. Full-grown rosettes of 5-week-old plants were used for experiments. All experiments (with the exception of the pulse labelling and fluorescence induction measurements in the presence of lincomycin) were conducted 2 h after the lights were turned on. For specific experiments, intact plants or detached leaves were treated at 900 μmol of photons·m−2·s−1 of white light (HL) as indicated. In search for the visual phenotype for ΔTLP18.3 plants, the plants were grown in 12 h dark/light rhythm, and fluctuating light was supplied from OSRAM Power Star HQIT metal halide lamps. Light intensity was adjusted with 1 h steps as follows: 15, 250, 350, 600, 250, 350, 600, 15, 600, 350, 250, 15 μmol of photons·m−2·s−1 by switching on and off the lamps.
Statistical analyses
The numerical data were subjected to statistical analysis by Student's t test, with statistical significance at the P values <0.001.
Isolation of polysomes
Polysomes were isolated from mature Arabidopsis leaves. The leaves were homogenized in 250 ml of grinding buffer (50 mM Hepes/KOH, pH 7.6, 330 mM sorbitol, 5 mM EDTA, 1 mM MgCl2, 5 mM ascorbate and 0.05% BSA), filtered through Miracloth and centrifuged at 1000 g at 4 °C for 1–2 min. The pellet was resuspended in the HSE buffer (50 mM Hepes/KOH, pH 8.0, 330 mM sorbitol and 2 mM EDTA) and loaded on to Percoll step gradients (70%: 3.5 ml of 100% Percoll and 1.5 ml of HSE; 40%: 6 ml of 100% Percoll and 9 ml of HSE). The Percoll step gradients were centrifuged at 4500 g at 4 °C for 5 min, and the lower part of the gradient containing intact chloroplasts was collected and diluted with 2–3-fold excess of the HSE buffer. Chloroplasts were pelleted at 1300 g at 4 °C for 2 min, resuspended in the HSE buffer and, after centrifugation at 1000 g at 4 °C for 1 min, again resuspended in the HSE buffer. Intact chloroplasts [equivalent to 250 μg of Chl (chlorophyll)] were lysed by adding 1 ml of lysis buffer containing a mixture of protease inhibitors (50 mM Hepes/KOH, pH 8.0, 5 mM magnesium acetate, 50 mM potassium acetate, 250 μg/μl chloramphenicol, 0.5 mg/ml heparin, 2 mM dithiothreitol, 100 μg/ml Pefabloc, 2 μg/ml antipain and 2 μg/ml leupeptin), and thylakoids were collected by centrifugation at 3500 g at 4 °C for 1 min. The lysis step was repeated twice. Thylakoids were solubilized with 1% (w/v) dodecyl β-D-maltoside in the lysis buffer (0.5 mg of chl/ml), loaded on the top of the 500 μl sucrose cushion (1 M sucrose in the lysis buffer), and the polysomes were collected by centrifugation at 270000 g at 4 °C for 1 h. After centrifugation, the supernatant was removed carefully and the pellet, if not used immediately, was promptly frozen by liquid nitrogen and stored at −80 °C.
In vivo pulse labelling of thylakoid proteins
Radioactive methionine was incorporated into detached leaves through petioles overnight (20 μCi·ml−1 [35S]methionine and 0.4% Tween 20). Subsequently, the pulse labelling was performed under HL conditions at 23 °C, and the samples were collected after 15, 30 and 60 min. For chase experiments, the leaves were washed with unlabelled methionine (10 mM L-methionine and 0.4% Tween 20), followed by 1 h incorporation of unlabelled methionine under darkness. Chase experiments were performed under HL conditions at 23 °C and the samples were collected after 1, 2 and 3 h. The rate constant for D1 degradation and the relative rates for D1 protein synthesis were calculated from the amount of radioactivity incorporated into D1 relative to that which was stably incorporated into CF1 (α- and β- subunits of the ATP synthase) under HL conditions and remained unchanged during the chase period. The relative synthesis rate of the D1 protein was determined from the shortest 15 min pulse samples, since the use of 30 min and 60 min pulses appeared to be hampered by the degradation of the newly labelled D1 protein (rapid D1 turnover at HL).
Isolation and fractionation of thylakoid membranes
Total thylakoids were isolated as described in [27]. Chl was determined according to the method described previously[28], and Chl per leaf area was calculated as described in [29]. For subfractionation, leaves were collected from GL conditions, and after HL treatment of plants for 6 days, thylakoids were isolated, diluted in the suspension buffer (50 mM Hepes/NaOH, pH 7.5, 10 mM MgCl2 and 100 mM sorbitol) to a Chl concentration of 0.2 μg/μl and 0.2% digitonin was added. The samples were vortex mixed for 4 s at room temperature. After centrifugation at 1000 g at 4 °C for 3 min, the supernatant was collected and centrifuged at 29000 g at 4 °C for 30 min to precipitate the grana membranes. To collect the stroma-exposed membranes, the supernatant was centrifuged at 185000 g for 4 °C for 90 min.
2D IEF (two-dimensional isoelectric focusing)
Polysomal proteins were separated by IEF in the first dimension followed by SDS/PAGE into the second dimension. Protein concentrations were determined by Lowry assay (BioRad). Proteins (100 μg) were solubilized in the RB-buffer [8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 100 mM dithiothreitol, 0.5% (v/v) Biolytes, pH 3–10] and then applied during 12 h of active rehydration to ReadyStrip™ IPG Strips pH 4–7 (BioRad). The proteins were focused using a programme: 150 V for 15 min, 300 V for 40 min, 500 V for 1 h, 1000 V for 2 h, followed by 8000 V until 65000 V/h was reached. After focusing the strips were incubated in 2% (w/v) dithiothreitol in EB buffer [6 M urea, 0.375 M Tris/HCl, pH 8.8, 2% (w/v) SDS, 20% (v/v) glycerol] at room temperature for 15 min, followed by incubation in 2.5% (w/v) iodoacetamide in EB buffer at room temperature for 15 min in the dark. Subsequently the strips were subjected to denaturing electrophoresis in the second dimension (SDS/PAGE, 14% gels) [30]. After electrophoresis, the proteins were visualized by silver staining [31].
BN (blue-native) gels
BN-PAGE was performed as described previously [32]. Thylakoid membranes were first resuspended in the 25BTH20G buffer [25 mM BisTris/HCl, pH 7.0, 20% (w/v) glycerol and 0.25 mg/ml Pefabloc] to a final Chl concentration of 0.5 mg/ml. An equal volume of 2% (w/v) dodecyl β-D-maltoside was added and the thylakoids were solubilized on ice for 3 min. Traces of unsolubilized material was removed by centrifugation at 18000 g at 4 °C for 20 min. Supernatant was supplemented with 1/10 volume of 100 mM BisTris/HCl (pH 7.0), 0.5 M ϵ-amino-n-hexanoic acid, 30% (w/v) sucrose and 50 mg/ml Serva Blue G. Proteins were loaded on BN-PAGE (5–12.5% acrylamide gradient) in amounts corresponding to 4 μg of Chl per well. Electrophoresis (Hoefer Mighty Small, Amersham Pharmacia Biotech, Uppsala, Sweden) was performed at 0 °C by gradually increasing the voltages as follows: 50 V for 30 min, 75 V for 30 min, 100 V for 1 h, 125 V for 30 min, 150 V for 1 h, 175 V for 30 min, followed by 200 V until the stain reached the end of the gel. After electrophoresis the lanes were excised and solubilized using the Laemmli buffer [30] containing 5% (v/v) 2-mercaptoethanol for 1 h at room temperature. The strips were run in the second dimension by SDS/PAGE (15% polyacrylamide and 6 M urea).
SDS/PAGE, immunoblotting and protein quantification
The polypeptides were separated by SDS/PAGE (15% polyacrylamide and 6 M urea) [30]. After electrophoresis, the proteins were electroblotted on to a PVDF membrane (Millipore, Watford, Herts., U.K.) and Western blotting with chemiluminescence detection was performed according to standard procedures using protein-specific antibodies or the antibody raised against the PSI complex. The TLP18.3 antibody was produced against synthetic peptides: DGQPDPGGPTVKDSK and RESNFKTKEETDEKR (Eurogentec, Seraing, Belgium). Lhcb1, Lhcb4 and PsbS antibodies were purchased from Agrisera (Vännäs, Sweden), and the antibody raised against the D1 protein was produced against oligopeptides as described previously [14]. The CP43 and PsbO antibodies were kindly provided by Dr R. Barbato, and the PsbP and PsbQ antibodies were kindly provided by Dr C. Spetea and Dr T. Hundal respectively. [35S]Methionine-labelled proteins were detected on PVDF membranes by autoradiography (X-ray film). Quantification of different proteins was carried out by densitometry analysis of the films with FluorChem™ 8000 image analyser (Alpha Innotech Corporation, San Leandro, CA, U.S.A.).
In-gel trypsin digestion and sample preparation for MS
In-gel trypsin digestion and sample preparation for MS were performed as described previously [32]. MALDI–TOF (matrix-assisted laser-desorption-ionization time-of-flight) analysis was performed in reflector mode on a Voyager-DE PRO mass spectrometer (Applied Biosystems). ESI (electrospray ionization)-MS was performed on a hybrid mass spectrometer API QSTAR Pulsar i (Applied Biosystems) equipped with a nanoelectrospray ion source (MDS Protana).
Photosynthetic activity measurements
PSII photochemical efficiency was determined as a ratio of variable fluorescence (FV) to maximal fluorescence (FM) measured from intact leaves with Hansatech Plant Efficiency Analyzer after a 30 min incubation in the dark. In experiments with inhibition of translation, 1 mM lincomycin was incorporated into leaves through petioles overnight.
Non-photochemical quenching was determined using PAM-2000 Fluorometer (Walz, Effeltrich, Germany). Leaves were adapted to darkness for 30 min before the measurements were taken. Non-photochemical quenching was induced by exposing the leaves to actinic white light of 1000 μmol of photons·m−2·s−1 and was calculated as (FM−FM′)/FM′ (where FM′ is the maximal fluorescence in the illuminated state) [33].
Light-saturated rates of steady-state oxygen evolution from thylakoids were recorded using the Hansatech oxygen electrode fitted to the DW1 chamber (Hansatech, King's Lynn, U.K.) in the presence of 1 mM DMBQ (2,6-dimethyl-p-benzoquinone) as an artificial electron acceptor.
Flash-induced increase and subsequent relaxation of Chl fluorescence yield were measured by the double-modulation fluorometer (PSI Instruments, Brno, Czech Republic) in the 150 μs to 100 s time range as described before [34].
The redox change of P700 was monitored by absorbance at 810 minus 860 nm, using the PAM-Fluorometer PAM-101/102/103 (Walz, Germany) equipped with the ED-P700DW-E emitter-detector unit. P700 was oxidized by far-red light from a photodiode (FR-102, Walz, Germany) for 30 s, and the subsequent re-reduction of P700+ in darkness was monitored.
RESULTS
Novel proteins associated with the polysomes
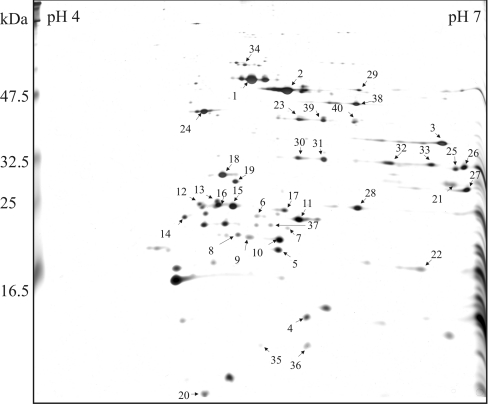
We have set up a proteomic system to search for novel thylakoid-associated regulatory proteins from the polysomes. A total of 40 proteins were identified by MS (Figure 1 and Table 1), most of which were subunits of the photosynthetic complexes or the translation apparatus, as expected. Subunits α, β, γ and ϵ of ATP synthase, as well as NdhH protein of NDH-1 complex, cytF and Rieske FeS proteins of cyt b6f complex, and PsbE, PsbO1 and PsbO2 subunits of PSII were observed. LHC (light-harvesting complex) proteins were also abundant: Lhca1, Lhca3, Lhca4 and Lhca5 proteins of LHCI, and Lhcb1, Lhcb3 and Lhcb5 proteins of LHCII complex, were identified. FNR (ferredoxin–NADP+ reductase) and FNR-like protein were detected as two spots having equal molecular masses, but different isoelectric points, possibly representing phosphorylated and dephosphorylated forms of the proteins. The detection of soluble FNR and FNR-like proteins corroborates earlier studies showing that a pool of FNR proteins are associated with Cyt b6f [35], NDH-1 [36] and PSI complexes [37]. Rubisco large subunit, being the most abundant stromal product of chloroplast protein synthesis, was not present as a contamination, but most probably represented proteins under translation process [38]. For translation apparatus, S1, S5 and L4 ribosomal proteins, as well as PSRP-2, representing a ribosomal regulatory protein, were found. In addition, the FtsH protease Var2 and four potential auxiliary proteins were identified: TLP18.3 and At1g15980 proteins found as one isoform, and the PsbQ domain protein At1g14150 and At1g64770 protein, both present in two isoforms. Of these proteins TLP18.3 was selected for further analysis.
Figure 1. Silver-stained gel after 2D IEF–SDS/PAGE separation of proteins from Arabidopsis thylakoid associated polysomes.
The polysomes were isolated from WT Arabidopsis plants grown under standard conditions; proteins were separated by 2D electrophoresis with denaturing IEF on immobilized pH gradient between 4 and 7 in the first dimension and with SDS/PAGE in the second dimension. The protein spots were identified with MALDI–TOF (identified proteins are listed in Table 1).
Table 1. Arabidopsis proteins identified from polysomes.
Mass spectrometric identification of Arabidopsis proteins isolated from chloroplast polysomes, as indicated in Figure 1. MM, molecular mass.
| Spot no. | Protein | AGI* | MM (kDa) | Peptides matched/total | Cover (%) | Score† |
|---|---|---|---|---|---|---|
| 1 | ATP synthase α subunit (AtpA) | ArthCp007 | 55.3 | 17/21 | 39 | 302 |
| 2 | ATP synthase β subunit (AtpB) | ArthCp029 | 54.0 | 21/26 | 69 | 359 |
| 3 | ATP synthase γ chain (AtpC1) | At4g04640 | 33.5 | 11/15 | 38 | 161 |
| 4 | ATP synthase ϵ chain (AtpE) | ArthCp028 | 14.5 | 4/6 | 38 | 83 |
| 5 | ATP synthase δ (OSCP) chain, putative H+-transporting ATPase | At4g09650 | 25.6 | 6/8 | 29 | 113 |
| 6 | PS I type III chlorophyll a/b binding protein (lhca3) | At1g61520 | 29.1 | 6/10 | 27 | 91 |
| 7 | PSI LHC protein Lhca5 | At1g45474 | 27.8p | 4/7 | 19 | 85 |
| 8 | PSI LHC protein Lhca4 | At3g47470 | 27.7p | 4/5 | 23 | 90 |
| 9 | PSI LHC protein Lhca1 | At3g54890 | 26.1 | 6/11 | 18 | 78 |
| 10 | PSI LHC protein Lhca1 | At3g54890 | 26.1 | 8/11 | 25 | 109 |
| 11 | PS I type III chlorophyll a/b-binding protein (Lhca3) | At1g61520 | 29.1 | 8/14 | 28 | 110 |
| 12 | Lhcb1 protein | At2g34430 | 28.2 | 8/15 | 44 | 107 |
| At2g34420 | ||||||
| 13 | Lhcb1 protein | At2g34430 | 28.2 | 8/12 | 44 | 124 |
| At2g34420 | ||||||
| 14 | Lhcb3 protein | At5g54270 | 28.7 | 7/9 | 43 | 102 |
| 15 | Lhcb1 protein | At1g29920 | 28.2 | 8/13 | 63 | 125 |
| At1g29910 | ||||||
| At1g29930 | ||||||
| 16 | Lhcb1 protein | At1g29920 | 28.2 | 7/13 | 47 | 98 |
| At1g29910 | ||||||
| At1g29930 | ||||||
| 17 | CP26 antenna protein of PS II (Lhcb5) | At4g10340 | 30.2 | 10/15 | 41 | 131 |
| 18 | 33 kDa oxygen-evolving enhancer protein (PsbO1) | At5g66570 | 35.3 | 12/15 | 43 | 221 |
| 19 | 33 kDa oxygen-evolving enhancer protein (PsbO2) | At3g50820 | 35.2 | 10/14 | 44 | 169 |
| 20 | Cytochrome B559 α chain (PsbE) | ArthCp039 | 9.3 | 5/7 | 28 | 85 |
| 21 | Cytochrome F (PetA) | ArthCp035 | 35.5 | 8/12 | 38 | 155 |
| 22 | Rieske FeS protein (PetC) | At4g03280 | 22.8 | 5/6 | 29 | 115 |
| 23 | NADH dehydrogenase, subunit 7 | ArthCp080 | 45.6 | 16/19 | 55 | 252 |
| 24 | 30 S ribosomal protein S1 | At5g29771 | 45.3 | 15/18 | 35 | 223 |
| 25 | 30 S ribosomal protein S5 | At2g33800 | 32.7 | 6/11 | 18 | 65 |
| 26 | 30 S ribosomal protein S5 | At2g33800 | 32.7 | 10/19 | 26 | 89 |
| 27 | 50 S ribosomal protein L4 | At1g07320 | 30.1 | 10/18 | 41 | 137 |
| 28 | RNA recognition motif (RRM)-containing protein (PSRP-2) | At3g52150 | 27.7 | 10/16 | 18 | 95 |
| 29 | Rubisco large subunit (RbcL) | ArthCp030 | 48.0 | 13/19 | 31 | 182 |
| 30 | FNR | At5g66190 | 40.6 | 10/15 | 33 | 119 |
| 31 | FNR | At5g66190 | 40.6 | 17/22 | 42 | 204 |
| 32 | FNR-like | At1g20020 | 39.1 | 15/19 | 37 | 214 |
| 33 | FNR-like | At1g20020 | 39.1 | 13/20 | 25 | 137 |
| 34 | FtsH protease (Var2) | At2g30950 | 74.3 | 14/19 | 29 | 169 |
| 35 | PsbQ domain protein family | At1g14150 | 22.2 | 4/8 | 21 | 64 |
| 36 | PsbQ domain protein family | At1g14150 | 22.2 | 5/7 | 31 | 113 |
| 37 | Thylakoid lumen 18.3 kDa protein (TLP18.3) | At1g54780 | 31.1 | 7/12 | 17 | 115 |
| 38 | Expressed protein At1g15980 | At1g15980 | 51.3 | 11/14 | 29 | 184 |
| 39 | Expressed unknown protein At1g64770 | At1g64770 | 38.3 | 7/11 | 24 | 123 |
| 40 | Expressed unknown protein At1g64770 | At1g64770 | 38.3 | 5/7 | 16 | 79 |
* Coding system of Arabidopsis protein coding genes, RNA coding genes and pseudogenes in MIPS (Munich Information Centre for Protein Sequences).
† Mascot protein score is −10×log(P), where P is the probability that the observed match is a random event.
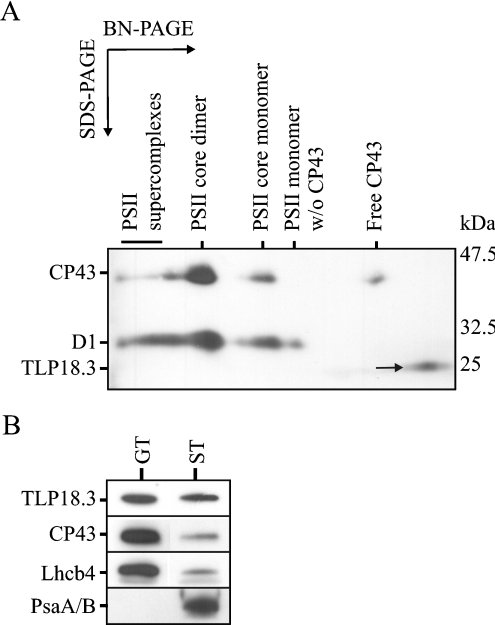
The TLP18.3 protein was found enriched in association with the polysomes, whereas lower amount of TLP18.3 was found from the total thylakoid preparation (results not shown). To clarify whether the TLP18.3 protein is a tightly bound subunit of some thylakoid protein complex, the thylakoids from WT Arabidopsis were subjected to 2D BN-PAGE analysis, enabling the separation of the protein complexes according to their size. Immunoblot analysis of the 2D BN-PAGE gel using antibodies raised against the D1, CP43 and TLP18.3 proteins showed that the TLP18.3 protein after BN-PAGE was not associated with any thylakoid protein complex, but instead migrated in the gel as a free protein (Figure 2A). The migration of the protein was highly dependent on the composition of the SDS/PAGE gel: if the separation of the polypeptides was carried out using a traditional SDS/PAGE gel without urea, the TLP18.3 protein migrated as an 18 kDa protein (37 in Figure 1), whereas when the peptides were separated by SDS/PAGE in the presence of 6 M urea, the protein migrated as a 25 kDa protein (Figure 2A).
Figure 2. Localization of the TLP18.3 protein in Arabidopsis thylakoids.
(A) Thylakoid membranes isolated from WT plants were subjected to 2D BN-PAGE; proteins were transferred to a PVDF membrane, followed by the analysis with antibodies against D1, CP43 and TLP18.3. (B) Thylakoid membranes were fractionated into stroma (ST) and grana (GT) thylakoids by digitonin, and proteins were separated by SDS/PAGE, followed by transfer to PVDF membrane and analysis with antibodies against TLP18.3, CP43, Lhcb4 and PsaA/B.
Localization of the TLP18.3 protein
In order to localize the lumenal TLP18.3 protein more precisely, WT thylakoids isolated from growth conditions were fractionated into the stroma lamellae and grana membranes by digitonin. The purity of the fractions was evaluated using the PSII core protein CP43, antenna protein Lhcb4 and the PSI core proteins PSIA/B as controls. As expected, the PSI core proteins were observed exclusively in the stroma-exposed thylakoids and the PSII proteins in the grana thylakoids, except for a small amount of PSII proteins in the stroma-exposed membrane representing the PSII complexes under repair (Figure 2B). Interestingly, the TLP18.3 protein was found more evenly distributed between the grana stacks and the stroma thylakoids, indicating that the protein might have a function in both locations (Figure 2B).
Screening of the ΔTLP18.3 T-DNA insertion mutants
To address the function of the TLP18.3 protein, the T-DNA insertion mutants were obtained from SALK and GABI-Kat collections (SALK_109618; GABI-Kat 459D12). The SALK_109618 line plants had the T-DNA insert in the first intron of the At1g54780 gene (see Supplementary Figure 1A at http://www.BiochemJ.org/bj/406/bj4060415add.htm), and in the GABI-Kat line 459D12 the T-DNA insert was located in the beginning of the last exon of the At1g54780 gene, between predicted domain of unknown function 477 (DUF477) and a transmembrane helix (TMH) (see Supplementary Figures 1A and 1B). SALK_109618 and GABI-Kat 459D12 plants selected for experiments were homozygous according to PCR analysis (data not shown) and did not contain detectable amounts of the TLP18.3 protein in thylakoids (see Supplementary Figure 1C).
Characterization of ΔTLP18.3 plants from normal growth conditions
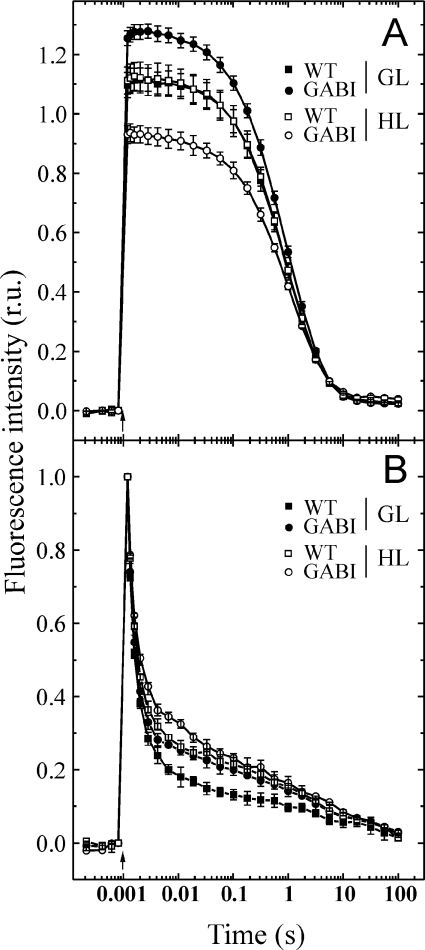
Arabidopsis plants lacking the TLP18.3 protein did not reveal any visible phenotype under standard growth conditions (results not shown). Chl a/b ratio, as well as the total amount of Chl per leaf area, were similar in WT and mutant plants (Table 2A). Activity of the PSI complex, monitored in vivo as redox changes of P700, the primary electron donor of the PSI complex, did not reveal any significant differences between WT and mutant plants (results not shown). However, the oxygen evolution activity of PSII was slightly higher in the mutant thylakoids compared with WT (Table 2A). To investigate whether the lack of the TLP18.3 protein has an influence on the acceptor and/or donor side of PSII, flash-induced increase and subsequent relaxation of fluorescence yield in the presence and absence of DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] was performed. The results showed slightly slower relaxation kinetics in mutant thylakoids compared with WT in the absence of DCMU (Figure 3B), demonstrating modifications on the acceptor side of the PSII complex. Interestingly, in the presence of DCMU, the overall fluorescence relaxation kinetics was identical in the WT and ΔTLP18.3 thylakoids. However, the total amplitude of fluorescence in the presence of DCMU, reflecting the amount of active PSII centres, was around 8% higher in ΔTLP18.3 thylakoids (Figure 3A). No statistically significant differences were recorded in the induction of non-photochemical quenching between WT and ΔTLP18.3 plants (results not shown).
Table 2. Characteristics of the photosynthetic apparatus in ΔTLP18.3 plants (GABI-Kat 459D12, SALK_109618).
Chl a/b ratio, total Chl/leaf area (μg Chl/cm2) and oxygen evolution capacity (μmol of O2/mg of Chl per h) of WT and ΔTLP18.3 plants. (A) Measurements from steady-state GL conditions (150 μmol of photons·m−2s−1). (B) After 3 h of mild HL treatment (900 μmol of photons·m−2·s−1). The HL values are presented as percentage value derived from the corresponding GL value. The results are means±S.D. (n=7 for Chl measurements and n=3–6 for oxygen evolution measurements).
| (A) Growth light | ||
|---|---|---|
| Parameter | WT | ΔTLP18.3 |
| Chl a/b | 3.2±0.02 | 3.1±0.09 (GABI-Kat) |
| 3.1±0.06 (SALK) | ||
| Chl/leaf area | 22.3±1.2 | 21.1±0.7 (GABI-Kat) |
| 22.5±0.7 (SALK) | ||
| O2 evolution | 426±5 | 474±3 (GABI-Kat) |
| 486±5 (SALK) | ||
| (B) High light | ||
| Parameter | WT | ΔTLP18.3 |
| O2 evolution | 94%±2% | 80%±2% (GABI-Kat) 69%±2% (SALK) |
Figure 3. Flash-induced increase and subsequent relaxation of the fluorescence yield of WT and ΔTLP18.3 thylakoids.
Chl fluorescence relaxation curves in the presence (A) and the absence (B) of DCMU were recorded from WT (■) and ΔTLP18.3 (●) thylakoids isolated from the plants grown at GL conditions (150 μmol of photons·m−2·s−1) and WT (□) and ΔTLP18.3 (○) thylakoids isolated after 3 h of HL treatment (900 μmol of photons·m−2·s−1). The curves represent means±S.D. for three independent measurements. (B) Fluorescence traces were normalized to the same amplitude for easy comparison of the kinetics. GABI, GABI-Kat 459D12.
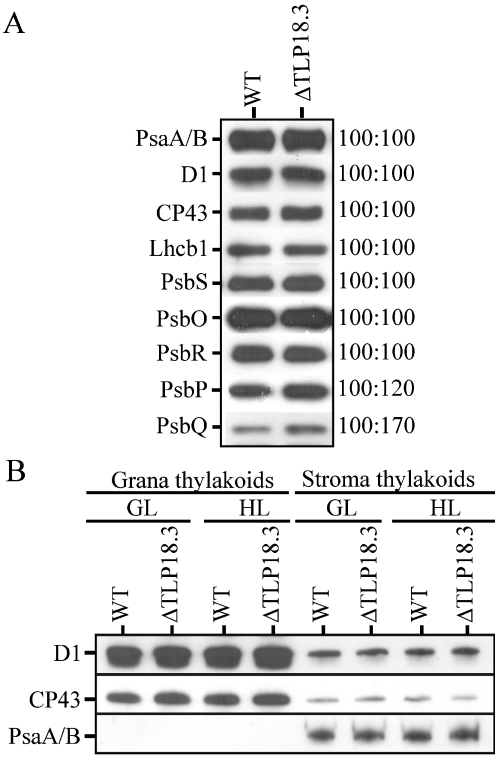
To analyse further the impact of the lack of the 18.3 kDa protein, an immunoblot analysis of thylakoid proteins was performed. Notable differences from WT were observed only in the PSII OEC (oxygen evolving complex) PsbQ protein and to a lesser extent in the PsbP protein, both being up-regulated in the ΔTLP18.3 plants (Figure 4A). The amounts of the PSI A/B proteins and the PSII core proteins D1 and CP43, as well as the PSII antenna proteins Lhcb1-6 (Lhcb2-6 results not shown) and the PsbS protein required for energy-dependent quenching, seemed to be unaltered in the mutant plants compared with WT plants (Figure 4A). It should be noted that the immunoblot analysis did not detect the minor increase in the quantity of the PSII complexes observed in the ΔTLP18.3 thylakoids with biophysical measurements.
Figure 4. Immunoblots of the thylakoid membrane proteins in WT and ΔTLP18.3 (GABI-Kat 459D12) mutant.
(A) Thylakoid membranes isolated from GL conditions; 0.25 μg of Chl was loaded, except for the detection of PsbS (0.5 μg), PsbR (0.5 μg), PsbP (0.5 μg) and PsbQ (2 μg), in order to maintain the amount of the protein in the linear range for the antibody used. A representative blot from three independent experiments is shown. On the right-hand side, the relative amount of protein in WT (given a value of 100) and mutant thylakoids is indicated. (B) Thylakoid membranes isolated from GL and HL conditions and fractionated into stroma and grana thylakoids by digitonin; 0.25 μg of Chl was loaded in order to maintain the amount of the protein in the linear range for the antibody used.
To test whether the distribution of the PSI and PSII complexes between grana and stroma membranes is affected by TLP18.3, an immunoblot analysis was performed using WT and GABI-Kat 459D12 thylakoids isolated from GL and HL, and subfractionated into grana and stroma membranes. As shown in Figure 4(B), the amount and distribution of the representative proteins of the PSII and PSI complexes were unaltered under both conditions.
Susceptibility of ΔTLP18.3 plants to short-term HL illumination
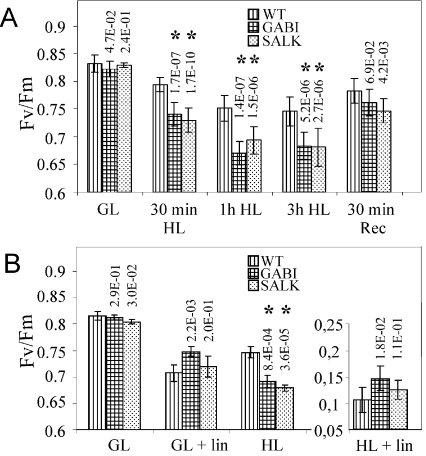
We next investigated the impact of the TLP18.3 protein in the susceptibility of the PSII activity to mild HL stress of 900 μmol of photons·m−2·s−1 by measuring the FV/FM fluorescence ratio from WT and ΔTLP18.3 plants. Under GL the FV/FM ratio for the WT and mutant plants was comparable (approx. 0.83; Figure 5A). A 30 min HL exposure caused a greater reduction in FV/FM for the GABI-Kat 459D12 (0.74) and SALK_109618 (0.75) plants relative to WT (0.79). HL treatment for 1 to 3 h further decreased the FV/FM ratio of WT plants to 0.75 and of the mutant plants close to 0.68 (Figure 5A). To study the mild HL stress-induced loss of PSII activity in the ΔTLP18.3 plants in more detail, the measurements of the steady-state oxygen evolution and the relaxation of flash-induced fluorescence yield were carried out. During 3 h of HL treatment the oxygen evolution activity of WT plants remained almost constant, while that of the ΔTLP18.3 plants diminished by 20% (459D12) or 30% (109618) (Table 2B). The flash-induced fluorescence relaxation measurements from thylakoids of HL-treated WT and ΔTLP18.3 plants showed slower fluorescence relaxation kinetics compared with GL thylakoids in the absence of DCMU (Figure 3B). However, in the ΔTLP18.3 thylakoids the overall fluorescence relaxation kinetics was more slowed down compared with WT, confirming modifications on acceptor side of PSII complex. No significant changes were observed in the fluorescence relaxation kinetics between HL-treated WT and ΔTLP18.3 plant thylakoids in the presence of DCMU (Figure 3A). Interestingly, the total amplitude of the flash-induced fluorescence in the presence of DCMU, reflecting the amount of active PSII centres, remained constant in WT thylakoids despite the HL treatment, whereas in the ΔTLP18.3 thylakoids the amplitude of the total fluorescence diminished by 20% during the 3 h HL treatment (Figure 3A). Both WT and ΔTLP18.3 plants were capable of rapid recovery after the HL treatment (Figure 5A). These results provided evidence that the deletion of the TLP18.3 protein led to enhanced photoinhibition of the PSII complex even under a mild HL illumination.
Figure 5. Changes in the photochemical efficiency of PSII (FV/FM) in WT and ΔTLP18.3 plants during HL treatment.
(A) Short-term HL treatment. Measurements were carried out using full-grown plants from GL (150 μmol of photons·m−2·s−1) 2 h after the lights were turned on. Plants were then transferred to HL (900 μmol of photons·m−2·s−1) and samples were taken after 30 min, 1 h and 3 h of illumination. Thereafter plants were transferred back to GL and the samples were taken after 30 min (Rec). The values are the means±S.D. (n=20). (B) HL treatment in the presence of lincomycin. Lincomycin was incorporated into detached leaves trough petioles overnight in darkness. Leaves floating on water, with or without 1 mM lincomycin, were exposed to GL (150 μmol of photons·m−2·s−1) or HL (900 μmol of photons·m−2·s−1) at 23 °C and the PSII activity was measured after 2 h of light treatment. The values are the means±S.D. (n=6). GABI, GABI-Kat 459D12; SALK, SALK_109618. The P values from experiments comparing the photoinhibition of the mutant plants with those of the corresponding WT plants are indicated on the top of the mutant columns, and statistically significant differences from WT are marked with asterisk (*). It should be noted that there were no statistically significant differences between the two TLP18.3 mutant lines in their susceptibility to photoinhibition (results not shown).
To analyse whether the enhanced PSII photoinhibition in ΔTLP18.3 plants is related to impaired repair of damaged PSII proteins via protein synthesis, the fluorescence induction measurements upon HL exposure were repeated in the presence of lincomycin (1 mM), a chloroplast translation inhibitor. WT, GABI-Kat 459D12 and SALK_109618 leaves were floated on water in a Petri dish in the presence or absence of lincomycin and exposed to either GL or HL for 2 h, followed by the PSII activity measurements. The water controls showed a similar photoinhibition pattern as obtained with leaves from intact plants, but, interestingly, in the presence of lincomycin the mutant plants seemed to be equally susceptible to photoinhibition as the WT plants (Figure 5B). It can thus be concluded that the higher susceptibility of ΔTLP18.3 plants to HL is due to ineffective PSII repair cycle.
Synthesis and degradation of the D1 protein in WT and ΔTLP18.3 plants at HL
To investigate whether HL-induced photoinhibition of ΔTLP18.3 plants was directly due to impaired de novo translation or degradation of the D1 protein, in vivo pulse–chase labelling experiments were performed. [35S]Methionine was incorporated into detached leaves through petioles in darkness overnight. Leaves were then pulse labelled under HL for 15, 30 and 60 min, followed by 1, 2 and 3 h chase in the presence of unlabelled methionine (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/406/bj4060415add.htm). The rate constants for D1 protein degradation were calculated by fitting the data to the first-order reaction kinetics. The rate constant for D1 protein degradation was significantly lower in the ΔTLP18.3 plants compared with WT plants (Table 3A). The apparent synthesis rate for D1 protein was calculated from the relative amount of radioactivity incorporated into the D1 protein during the 15 min pulse experiment. Like D1 degradation, the relative synthesis rate of the D1 protein was lower in mutant plants compared with WT, although the difference in synthesis was less pronounced than in the degradation (Table 3B). These results would indicate that the TLP18.3 protein had a direct role in the turnover of the D1 protein on the stroma-exposed thylakoids.
Table 3. Comparison of the rate constants (kPI) for D1 protein degradation and the relative synthesis rates of the D1 protein in the WT and ΔTLP18.3 thylakoids.
Detached leaves from WT and ΔTLP18.3 plants were pulse labelled under HL (900 μmol of photons·m−2·s−1) with [35S]methionine for 15, 30 and 60 min, followed by 1, 2 and 3 h chase in the presence of unlabelled methionine (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/406/bj4060415add.htm). (A) kPI and half-times (T½, min) for the D1 protein degradation were calculated from data fitted to first-order reaction kinetics (r=k[A]). Data shown are means±S.D., n=3 for WT and GABI-Kat 459D12 mutant and n=1 for SALK_109618 mutant. (B). Relative D1 protein synthesis rates were determined by comparing the amount of radioactivity incorporated into the D1 protein normalized to that incorporated into ATP synthase α-/β- subunits in WT and ΔTLP18.3 thylakoids during radioactive pulse of 15 min at HL. Data shown are means±S.D., n=3.
| (A) | ||
|---|---|---|
| Lines | kPI×103, min−1 | T½, min |
| WT | 8.1±1.1 | 87±13 |
| GABI-Kat 459D12 | 4.4±1.1 | 163±42 |
| SALK_109618 | 5.0 | 138 |
| (B) | ||
| Lines | Relative synthesis rate | |
| WT | 1.00±0.00 | |
| GABI-Kat 459D12 | 0.69±0.15 | |
| SALK_109618 | 0.69±0.05 |
Heterogeneity of the PSII complexes in WT and ΔTLP18.3 plants
To clarify the impact of the lack of the TLP18.3 protein for the organization of the PSII complexes, the relative amounts of the PSII dimers (PSII supercomplexes and PSII core dimers), PSII monomers and PSII monomers without CP43 subunit in the WT and ΔTLP18.3 thylakoids were quantified by 2D BN-PAGE combined with the immunoblot analysis of the D1 protein. Interestingly, under GL conditions the mutant plants had less of the PSII dimers and more of the PSII monomers, possibly under repair, than WT plants (Table 4). Moreover, although WT plants seemed to accelerate the PSII repair cycle under short-term HL treatment by increasing the proportion of the PSII monomers from 30% under GL to 36% under HL, the relative amount of PSII monomers in the ΔTLP18.3 plants remained at 45% despite the increase in light intensity (Table 4). This suggests that the ΔTLP18.3 plants were unable to alter their capacity to repair PSII upon increasing light intensity. Increasing instability of PSII dimers was seen in WT plants after HL shift, as shown by high standard deviations at HL, and was probably originating from general instability of the PSII complexes under photoinhibitory conditions. Such instability was evident for PSII dimers of the ΔTLP18.3 plants even under GL (Table 4).
Table 4. Quantification of the PSII complexes separated by 2D BN-PAGE (WT and GABI-Kat 459D12 thylakoids).
Relative amounts of the PSII dimers (PSII supercomplexes and PSII core dimers), PSII monomers and PSII monomers without CP43 subunit were defined using immunoblotting with D1 antibody, and results from three independent experiments were analysed. Thylakoids were isolated from steady-state GL conditions (150 μmol of photons·m−2·s−1) and after 3h mild HL treatment (900 μmol of photons·m−2·s−1) of plants.
| Relative amount (%) | ||
|---|---|---|
| PSII organization | WT | ΔTLP18.3 |
| GL | ||
| PSII dimers | 70±0.7 | 54±3.5 |
| PSII monomers | 25±2.1 | 36±3.5 |
| PSII monomers w/o CP43 | 5±2.8 | 10±0.1 |
| HL | ||
| PSII dimers | 64±7.8 | 56±7.9 |
| PSII monomers | 26±4.0 | 30±5.1 |
Retarded growth of the ΔTLP18.3 plants under fluctuating light
Since the ΔTLP18.3 plants showed no visible phenotype when grown under normal growth conditions (150 μmol of photons·m−2·s−1 in 8 h light/16 h dark regime) or at HL intensity (900 μmol of photons·m−2·s−1 in 8 h light/16 h dark regime), we decided to grow the plants under fluctuating light that requires strict regulation of light reactions. Relatively low irradiances were given in the beginning and end of the light period, whereas during midday the light intensity varied between 15 and 600 μmol of photons·m−2·s−1 (for the exact programming of the lamp, see the Experimental section). Under these conditions the growth of the ΔTLP18.3 plants was compromised as compared with WT (Figure 6).
Figure 6. Mutant phenotype.
The phenotype of WT, SALK_109618 and GABI-Kat 459D12 plants grown under fluctuating light conditions (see the Experimental section for details).
GABI-Kat 459D12 mutant plants accumulated a truncated form of TLP18.3 protein upon prolonged HL treatment
When studying the GABI-Kat 459D12 mutant thylakoids isolated after prolonged HL treatment of the plants, the antibody raised against the TLP18.3 protein recognized a protein approx. 2 kDa smaller than the TLP18.3 (Figure 7). This protein accumulated during HL treatment, in particular during the subsequent recovery at GL, while the content of the TLP18.3 protein in WT remained unchanged irrespective of the light condition (Figure 7). Interestingly, mass spectrometric identification revealed the protein as TLP18.3 (see Supplementary Figures 1D and 1E). Sequencing of the GABI-Kat 459D12 At1g54780 gene (results not shown) confirmed that the T-DNA was inserted into the gene after base pair 1841 of the unspliced At1g54780 gene, corresponding to amino acid 259 of the protein sequence. This led to the translation of a truncated TLP18.3 protein (termed TLP18.3*) where the last 26 amino acids from the C-terminus of the native protein were replaced with residues ITYIS (see Supplementary Figure 1D). Using protein domain prediction programs (TMHMM, SignalP, InterPro) the predicted TLP18.3* protein comprised the chloroplast-targeting signal, the DUF477 domain but not the C-terminal TMH sequence (see Supplementary Figures 1A and 1B). The absence of TLP18.3* in thylakoids isolated from the mutant plants grown under normal growth conditions is consistent with the requirement of the TMH sequence for the binding of TLP18.3 (albeit loosely, Figure 2A) to thylakoid membranes. However, after exposure to prolonged HL, increasing amounts of TLP18.3* peptides were isolated with the thylakoids from the GABI-Kat 459D12 mutant. The basis for this curious accumulation of TLP18.3* following HL treatment remains to be resolved.
Figure 7. Truncated form of the TLP18.3 protein (TLP18.3*) is expressed in GABI-Kat 459D12 plants.
WT and ΔTLP18.3 (GABI-Kat 459D12) plants growing under GL (150 μmol of photons·m−2·s−1) were transferred to HL (900 μmol of photons·m−2·s−1) for 5 h, 1 day, 3 days and 6 days. Thereafter plants were transferred back to GL (Rec) for 5 h and 1 day. Thylakoids were isolated, polypeptides separated by SDS/PAGE (1 μg of Chl per well) and an immunoblot analysis using the antibody raised against the TLP18.3 protein was carried out.
DISCUSSION
Proteome search for auxiliary PSII proteins
We have addressed the regulatory network related to PSII function and found potential novel photosynthetic regulatory proteins from thylakoids of Arabidopsis thaliana. During the past few years, hundreds of Arabidopsis chloroplast proteins have been localized in the inner envelope membrane, thylakoid membrane and thylakoid lumen with prediction programs and different proteomic approaches giving further insights into the function of unknown proteins [7–9,39]. In the present study, the IEF/SDS/PAGE gel system linked to mass spectrometric identification of proteins was used as a method to search unknown auxiliary proteins from polysomes likely to contain proteins involved directly or indirectly in the process of translation and assembly of chloroplast-encoded proteins (Figure 1 and Table 1). The method presented is especially promising in search for novel proteins involved in the synthesis of PSII subunits exhibiting a high turnover rate [24]. Enrichment in the polysome fraction of auxiliary proteins that generally are present only in minor amounts in the chloroplast was an additional benefit of using the polysomes for identification of novel regulatory proteins.
Thylakoid solubilization with dodecyl β-D-maltoside releases the ribosome nascent chain complexes associated with fragments of the stroma thylakoids, and indeed, stroma-exposed thylakoid proteins were strongly represented among identified proteins. Ribosomal proteins, on the other hand, were under-represented. The majority of ribosomal proteins are highly basic, having an isoelectric point in the pH range 9 to 10 [40], and therefore they were not separated by IPG strips selected for the search of novel auxiliary proteins in the pH range 4 to 7 (Figure 1). However, four ribosomal proteins were identified as well as one translation regulation protein, PSRP-2 (Table 1). PSRP-2 contains two RNA-binding domains, thus suggesting a function in recruiting of stored chloroplast mRNAs for active protein synthesis [41]. Interestingly, one known auxiliary protein, FtsH protease Var2, involved in the degradation of PSII D1 protein [12] was found enriched in polysomes as well as four auxiliary protein candidates: At1g14150 (a PsbQ family protein), At1g64770, At1g15980 and TLP18.3. The TLP18.3 protein, which was in earlier studies shown to be located in the thylakoid lumen of Arabidopsis [8,9], was selected for further analysis.
Analysis of the sequence of the TLP18.3 protein did not reveal any known functional domains. However, it is interesting to note that the TMH domain appears to play a role in the functional association of TLP18.3 with thylakoid membranes. As expected, the GABI-Kat 459D12 mutant plants showed no trace of the TLP18.3 protein, but did accumulate the C-terminally truncated TLP18.3* variant during prolonged HL treatment and the subsequent recovery period (Figure 7). According to our hypothesis the full-length TLP18.3 protein is loosely bound to the thylakoid membrane by the TMH, possibly in interaction with an unknown subunit of the PSII complex (see below), and thus the TLP18.3 protein is protected from degradation. This conclusion is based on two facts: firstly the TLP18.3 protein must be bound to the membrane, because it was detected in the polysomal preparation, which does not contain thylakoid lumenal proteins, and secondly the linkage of TLP18.3 to PSII is loose, because in the BN analysis the TLP18.3 protein was observed as a free protein (Figures 1 and 2A). Unlike the TLP18.3 protein, the truncated TLP18.3* protein produced by the GABI-Kat 459D12 plants is not bound to the membrane and is rapidly degraded under normal growth conditions (see Supplementary Figure 1C), but becomes more stabilized during prolonged HL stress, possibly due to the inactivation of the protease (Figure 7).
TLP18.3 is a novel lumenal protein required for high light tolerance of PSII
Characterization of the ΔTLP18.3 thylakoids from normal growth conditions revealed only small differences as compared with WT thylakoids. These include a higher amount of PSII reaction centres, as shown by the higher amplitude of the flash-induced fluorescence yield and by the higher steady-state oxygen evolution rate (Table 2A and Figure 3A), accompanied by an up-regulation of the PsbP and PsbQ proteins (Figure 4A). Despite only small differences under growth conditions, they are clear symptoms of disturbances in the light tolerance of the PSII complex [42]. Indeed, the physiological role of the TLP18.3 protein in protection against HL was revealed when plants were exposed to moderately high irradiances that enhance the PSII photoinhibition repair cycle. Observed higher susceptibility of the ΔTLP18.3 plants to HL can, in principle, originate from two sources; either the PSII complexes are more susceptible to light-induced damage, or the repair of the damaged PSII complexes is inefficient [43]. To differentiate between these two possibilities, the photoinhibition experiments were repeated in the presence of lincomycin to block the repair process. Now the enhanced susceptibility of ΔTLP18.3 plants to photoinhibition disappeared, indicating that the TLP18.3 protein facilitates the repair of the PSII centres after photodamage (Figure 5B). The repair of photodamaged PSII complexes is a complex process and occurs in multiple steps, including the monomerization and partial disassembly of the dimeric complex in grana stacks [5,44], migration of the PSII monomers to the stroma-exposed thylakoids [45,46], followed by proteolysis of the damaged D1 protein [5,13,47]. After co-translational insertion of the newly synthesized D1 protein into the membrane [14], the D1 protein is post-translationally processed, the PSII monomers are re-assembled [15,48] and migrate back to the grana thylakoids, where dimerization takes place.
Functional role of the TLP18.3 protein in PSII repair
It is important to specify the role of the TLP18.3 protein in the repair of PSII, due to the fact that the protein does not contain functionally known domains. It is also important to emphasize that the amount of the TLP18.3 protein is not regulated by light, but the content of the TLP18.3 protein remained constant in the thylakoid membrane regardless of the irradiance level (Figure 7), suggesting an intimate interaction with the PSII complex. The observed higher levels of PSII monomers in ΔTLP18.3 plants confirm that the monomerization of the PSII complex is not the limiting factor slowing down the repair process (Table 4). The migration of the PSII complexes from grana to stroma-exposed thylakoids does not seem to be a problem in the ΔTLP18.3 plants, since similar amounts of PSII complexes were recorded in the stroma thylakoids of both the WT and ΔTLP18.3 plants (Figure 4B). This latter conclusion is further supported by the fact that also cyanobacteria, which completely lack the thylakoid heterogeneity, have a homologue of TLP18.3, the Sll1390 protein. Sll1390 was found in sub-stoichiometric concentrations from active oxygen-evolving PSII complexes purified from Synechocystis sp. PCC 6803 using His-tagged CP47, strongly suggesting a role for SLL1390 as an auxiliary protein of PSII [49]. TLP18.3 thus seems to belong to evolutionarily conserved auxiliary proteins facilitating the assembly and repair of PSII, together with the previously characterized proteins Psb27 (slr1645) and Psb29 (Sll1414), both also found in purified Synechocystis PSII preparations [19,22,49,50].
Two steps in the repair process of PSII are clearly malfunctioning in the absence of the TLP18.3 protein, namely the turnover of the damaged D1 protein and the dimerization of the PSII complexes (Tables 3 and 4, and Supplementary Figure 2). Slower D1 degradation was measured from ΔTLP18.3 plants (Table 3A), and the enrichment of both the TLP18.3 protein and the D1-degrading protease FtsH in the same fraction further support the co-operating of these two proteins (Figure 1). Turnover of the photodamaged D1 protein in stroma-exposed thylakoid membranes precedes the dimerization of the PSII complex, and thus it would be logical to assign the function for the TLP18.3 protein solely in degradation and synthesis steps of D1 in stroma-exposed membranes. Nevertheless, the TLP18.3 protein was quite evenly distributed in both the grana stacks and stroma lamellae, suggesting a function also in the appressed membranes (Figure 2B). The increased amount of PSII monomers in TLP18.3 thylakoids as compared with WT thylakoids (Table 4) must reside in grana membranes, since the content of PSII centres was similar in grana and stroma thylakoids between WT and TLP18.3 (Figure 4B). Moreover, under growth conditions these complexes were clearly active (Figure 3A), indicating that they do not represent damaged PSII monomers, but active ones, apparently just after repair and migration back to the grana. Notably, the ΔTLP18.3 plants also exhibited increased amounts of the OEC proteins PsbP and PsbQ, which reflects a dysfunction of the dimerization of the PSII complex (Figure 4A). Indeed, it has been shown earlier that the assembly-competent OEC proteins can be stored into the thylakoid lumen for PSII repair [42,51,52] and that the PsbP and PsbQ proteins are bound not to stromal PSII monomers, but instead to the PSII dimers in the grana [32,52].
Taken together, our results provide evidence that the TLP18.3 protein is involved in regulation of both the degradation/synthesis steps of the damaged D1 protein of the PSII core in stroma-exposed membranes and (directly or indirectly) in the assembly of PSII monomers into dimers in grana stacks. HL illumination of the mutant plants, however, demonstrated that the PSII repair cycle does not completely collapse in the absence of the TLP18.3 protein, but rather the TLP18.3 protein represents an auxiliary protein that normally augments specific steps in the complicated processes of PSII repair. The beneficial role of TLP18.3 for optimal growth was particularly evident under fluctuating illumination, where the growth phenotype of the ΔTLP18.3 plants was compromised. This presumably resulted from perturbations in the capacity of PSII repair to adjust rapidly to varying rates of PSII damage during variations in illumination intensity.
Online data
Acknowledgments
This work was financially supported by the Academy of Finland, Biological Interactions Graduate School, Jenny and Antti Wihuri Foundation and Kone Foundation. The Proteomics and Mass Spectrometry Unit in the Turku Centre for Biotechnology is thanked for technical advices in protein analysis. We thank the Salk Institute Genomic Analysis Laboratory and GABI-Kat for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation and GABI-Kat by the German Plant Genomics Programme of BMBF.
References
- 1.Anderson J. M., Park Y., Chow W. S. Unifying model for the photoinactivation of photosystem II in vivo under steady-state photosynthesis. Photosynt. Res. 1998;56:1–13. [Google Scholar]
- 2.Asada K. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 3.Baena-Gonzalez E., Aro E. M. Biogenesis, assembly and turnover of photosystem II units. Philos.Trans. R. Soc. Lond. B. Biol. Sci. 2002;357:1451–1459. doi: 10.1098/rstb.2002.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber J., Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 5.Aro E. M., Virgin I., Andersson B. Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Soderberg L., Roepstorff P., von Heijne G., van Wijk K. J. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubert M., Petersson U. A., Haas B. J., Funk C., Schroder W. P., Kieselbach T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 10.Li X. P., Gilmore A. M., Niyogi K. K. Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 2002;277:33590–33597. doi: 10.1074/jbc.M204797200. [DOI] [PubMed] [Google Scholar]
- 11.Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., Jahns P., Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- 12.Bailey S., Thompson E., Nixon P. J., Horton P., Mullineaux C. W., Robinson C., Mann N. H. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- 13.Haussuhl K., Andersson B., Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 1999;274:16062–16067. doi: 10.1074/jbc.274.23.16062. [DOI] [PubMed] [Google Scholar]
- 15.Anbudurai P. R., Mor T. S., Ohad I., Shestakov S. V., Pakrasi H. B. The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8082–8086. doi: 10.1073/pnas.91.17.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danon A., Mayfield S. P. Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J. 1991;10:3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somanchi A., Barnes D., Mayfield S. P. A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J. 2005;42:341–352. doi: 10.1111/j.1365-313X.2005.02378.x. [DOI] [PubMed] [Google Scholar]
- 18.Yohn C. B., Cohen A., Rosch C., Kuchka M. R., Mayfield S. P. Translation of the chloroplast psbA mRNA requires the nuclear-encoded poly(A)-binding protein, RB47. J. Cell Biol. 1998;142:435–442. doi: 10.1083/jcb.142.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keren N., Ohkawa H., Welsh E. A., Liberton M., Pakrasi H. B. Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in synechocystis and arabidopsis. Plant Cell. 2005;17:2768–2781. doi: 10.1105/tpc.105.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. Low PSII Accumulation1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell. 2006;18:955–969. doi: 10.1105/tpc.105.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plucken H., Muller B., Grohmann D., Westhoff P., Eichacker L. A. The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 2002;532:85–90. doi: 10.1016/s0014-5793(02)03634-7. [DOI] [PubMed] [Google Scholar]
- 22.Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., Lu C., Zhang L. A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 2006;61:567–575. doi: 10.1007/s11103-006-0031-x. [DOI] [PubMed] [Google Scholar]
- 23.Falk H. Rough thylakoids: polysomes attached to chloroplast membranes. J. Cell Biol. 1969;42:582–587. doi: 10.1083/jcb.42.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattoo A. K., Hoffman-Falk H., Marder J. B., Edelman M. Regulation of protein metabolism: coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32 kDa protein of the chloroplast membranes. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1380–1384. doi: 10.1073/pnas.81.5.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 26.Rosso M. G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 27.Rintamaki E., Kettunen R., Aro E. M. Differential D1 dephosphorylation in functional and photodamaged photosystem II centers: dephosphorylation is a prerequisite for degradation of damaged D1. J. Biol. Chem. 1996;271:14870–14875. doi: 10.1074/jbc.271.25.14870. [DOI] [PubMed] [Google Scholar]
- 28.Porra R. J., Thompson W. A., Kriedemann P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and b with four different solvents: verification of the concentration of chlorophyll by atomic absorption spectroscopy. Biochim. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- 29.Inskeep W. P., Bloom P. R. Extinction coefficients of chlorophyll a and b in N, N-dimethylformamide and 80% acetone. Plant Physiol. 1985;77:483–485. doi: 10.1104/pp.77.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell K. L., Stults J. T. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997;18:349–359. doi: 10.1002/elps.1150180309. [DOI] [PubMed] [Google Scholar]
- 32.Rokka A., Suorsa M., Saleem A., Battchikova N., Aro E. M. Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem. J. 2005;388:159–168. doi: 10.1042/BJ20042098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilger W., Björkman O. Relationships among violaxanthin deepoxidation, thylakoid membrane conformation, and nonphotochemical chlorophyll fluorescence quenching in leaves of cotton (Gossypium hirsutum L.) Planta. 1994;193:238–246. [Google Scholar]
- 34.Allahverdiyeva Y., Deak Z., Szilard A., Diner B. A., Nixon P. J., Vass I. The function of D1-H332 in photosystem II electron transport studied by thermoluminescence and chlorophyll fluorescence in site-directed mutants of synechocystis 6803. Eur. J. Biochem. 2004;271:3523–3532. doi: 10.1111/j.0014-2956.2004.04287.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H., Whitelegge J. P., Cramer W. A. Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 2001;276:38159–38165. doi: 10.1074/jbc.M105454200. [DOI] [PubMed] [Google Scholar]
- 36.Guedeney G., Corneille S., Cuine S., Peltier G. Evidence for an association of ndh B: ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 1996;378:277–280. doi: 10.1016/0014-5793(95)01473-x. [DOI] [PubMed] [Google Scholar]
- 37.Andersen B., Scheller H. V., Moller B. L. The PSI-E subunit of photosystem I binds ferredoxin:NADP+ oxidoreductase. FEBS Lett. 1992;311:169–173. doi: 10.1016/0014-5793(92)81391-x. [DOI] [PubMed] [Google Scholar]
- 38.Muhlbauer S. K., Eichacker L. A. The stromal protein large subunit of ribulose-1,5-bisphosphate carboxylase is translated by membrane-bound ribosomes. Eur. J. Biochem. 1999;261:784–788. doi: 10.1046/j.1432-1327.1999.00337.x. [DOI] [PubMed] [Google Scholar]
- 39.Ferro M., Salvi D., Riviere-Rolland H., Vermat T., Seigneurin-Berny D., Grunwald D., Garin J., Joyard J., Rolland N. Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11487–11492. doi: 10.1073/pnas.172390399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaltschmidt E., Wittmann H. G. Ribosomal proteins. XII. number of proteins in small and large ribosomal subunits of Escherichia coli as determined by two-dimensional gel electrophoresis. Proc. Natl. Acad. Sci. U.S.A. 1970;67:1276–1282. doi: 10.1073/pnas.67.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi K., Subramanian A. R. Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur. J. Biochem. 2003;270:190–205. doi: 10.1046/j.1432-1033.2003.03359.x. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto A., Yamamoto Y., Theg S. M. Unassembled subunits of the photosynthetic oxygen-evolving complex present in the thylakoid lumen are long-lived and assembly-competent. FEBS Lett. 1996;391:29–34. doi: 10.1016/0014-5793(96)00686-2. [DOI] [PubMed] [Google Scholar]
- 43.Adir N., Zer H., Shochat S., Ohad I. Photoinhibition: a historical perspective. Photosynth. Res. 2003;76:343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- 44.Hankamer B., Barber J., Boekema E. J. Structure and membrane organization of photosystem II in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:641–671. doi: 10.1146/annurev.arplant.48.1.641. [DOI] [PubMed] [Google Scholar]
- 45.Aro E. M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamaki E. Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 2005;56:347–356. doi: 10.1093/jxb/eri041. [DOI] [PubMed] [Google Scholar]
- 46.van Wijk K. J., Andersson B., Aro E. M. Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 1996;271:9627–9636. doi: 10.1074/jbc.271.16.9627. [DOI] [PubMed] [Google Scholar]
- 47.Lindahl M., Tabak S., Cseke L., Pichersky E., Andersson B., Adam Z. Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 1996;271:29329–29334. doi: 10.1074/jbc.271.46.29329. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell. 2000;12:1769–1782. doi: 10.1105/tpc.12.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–8012. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- 50.Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rogner M. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ettinger W. F., Theg S. M. Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J. Cell Biol. 1991;115:321–328. doi: 10.1083/jcb.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto A., Ettinger W. F., Yamamoto Y., Theg S. M. Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: sites of assembly and mechanism of binding. Plant Cell. 1997;9:441–452. doi: 10.1105/tpc.9.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.