Abstract
Classic galactosemia is an autosomal recessive inherited error of galactose metabolism. It is caused by lack of galactose-1-phosphate uridyl transferase, an enzyme that is required to metabolize galactose-1-phosphate to uridine diphosphate galactose. The build up of galactose-1-phosphate is toxic at high levels and can damage the liver, brain, eyes, and other vital organs. Over 200 mutations have been identified in affected individuals. We describe an assay to identify nine target mutations or variants in the galactose-1-phosphate uridyl transferase gene, namely p.Q188R, p.S135L, p.K285N, p.L195P, p.T138M, p.Y209C, IVS2-2 A>G, p.L218L, and p.N314D. A single long-range PCR is followed by a multiplexed nucleotide extension assay (single nucleotide extension) and capillary electrophoresis to detect simultaneously all nine target mutations/variants. Fifty-four previously characterized samples (47 clinical samples and seven controls) gave a 100% concordance. We also report a nontarget novel mutation, p.L192X, and its profile using single nucleotide extension. This assay can complement the enzyme activity assay and identify familial mutations for testing additional family members.
Classic galactosemia is an inherited metabolic disorder caused by a deficiency of galactose-1-phosphate uridyl transferase (GALT), an enzyme required to convert galactose-1-phosphate to uridine diphosphate galactose.1,2,3,4 The resulting buildup of galactose-1-phosphate leads to poor feeding, vomiting, diarrhea, and cataracts, and if left untreated, results in physical and mental retardation.5,6,7,8 Over 200 mutations have been identified in patients affected with galactosemia (http://arup.utah.edu/database/galactosemia/GALT_welcome.php). The six common mutations p.Q188R (c.563A>G), p.S135L (c.404 C>T), p.K285N (c.855G>T), p.L195P (c.584T>C), p.T138M (c.413 C>T), and p.Y209C (c.626 A>G) account for 80% of cases with classic galactosemia. A seventh mutation, IVS2-2 A>G (c.253-2A>G), has been described in a Hispanic population.9 These mutations are considered severe or classic mutations resulting in little or no enzyme activity. Two variants, Los Angeles (LA or Duarte 1) and Duarte, also affect enzyme activity. The LA variant p.L218L (c.652C>T) in cis with the p.N314D (c.940A>G) increases the enzyme activity to approximately 130%.10 The Duarte (D) variant (p.N314D, c.940A>G) reduces enzyme activity by approximately 25% and is found in approximately 5% of the general US population.11,12 This variant is also in cis with the deletion c.-119->-116delGTCA in the 5′-untranslated region of the GALT gene, thus also affecting a regulatory region.13,14 The combination of a classic mutation (G) and the D variant shows a decreased enzyme activity but does not typically result in symptoms of classic galactosemia.
Galactosemia screening is widely performed on newborns. Blood specimens are collected on Guthrie cards and assayed for galactose and its metabolite, galactose 1-phosphate, or GALT enzyme levels.3,5,15,16 Abnormal screening tests are followed by confirmatory testing using enzymatic activity.17 The GALT enzyme is unstable, and the assay for enzyme activity is best performed on fresh heparin anticoagulated blood. DNA analysis for common mutations can be used in conjunction with enzyme activity to confirm familial mutations. Our purpose was to design a practical method to detect the seven GALT mutations and the two variants. We chose the mutations and variants based on a Texas newborn population study.9 The IVS2-2 mutation was included to increase detection in Hispanic populations. PCR primers were designed to produce a single amplicon that included all mutation and variant positions followed by multiplexed single nucleotide extension (SNE).
Materials and Methods
Samples
As part of the State of Utah newborn galactosemia screening, newborns with moderate to high elevated levels of galactose, defined as 20 to 30 mg/dL, were sent for confirmatory enzyme activity levels and DNA mutation analysis. DNA was extracted by the MagNaPure instrument (Roche, Indianapolis, IN). DNA from 47 de-identified clinical samples previously characterized by ARUP Laboratories (by enzyme activity and/or allele-specific PCR) was tested following an Institutional Review Board-approved protocol for test validation. Six additional blinded DNA samples previously characterized by the Department of Genetics, Emory University School of Medicine, and known to contain seven mutations and one variant were also tested. An artificial template was synthesized for IVS 2-2A>G heterozygous mutation, for a total of 54 samples.
Artificial Template for IVS2-2 A>G
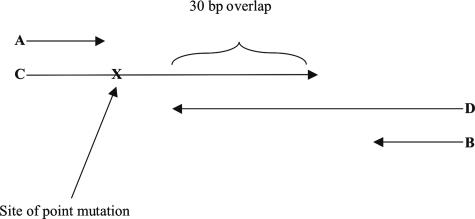
To generate the artificial template for IVS2-2 A>G mutation18,19,20 (Figure 1 and Table 1), two oligodeoxyribonucleotides of 90 and 91 bases that overlapped by 30 bp were used to introduce the IVS2-2 G mutation. In addition, two short primers (IVS2-2 forward and IVS2-2 reverse) were used in the same reaction. This allowed generation of the template by overlapping oligodeoxyribonucleotides followed by exponential amplification with the short primers. A 20-μl PCR was performed with 0.5 μmol/L of each of the four oligodeoxyribonucleotides, 1× FailSafe Premix D (Epicenter, Madison, WI), and 1.0 unit of FastStart DNA polymerase (Roche). Amplification was performed with an initial denaturation at 95°C for 5 minutes, followed by five cycles of denaturation at 94°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 30 seconds to generate the template. This was followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. Unincorporated primers and dNTPs were removed by incubating with ExoSAP (USB Corporation, Cleveland, OH) as described below. To mimic a heterozygous sample, the purified artificial template was mixed with a wild-type genomic DNA.
Figure 1.
Schematic of primer locations for the generation of artificial template for IVS 2-2 A>G mutation. A: IVS 2-2 forward primer; B: IVS 2-2 reverse primer; C: artificial oligodeoxyribonucleotides template forward primer; and D: artificial oligodeoxyribonucleotides template reverse primer. “X” indicates the position of the mutation.
Table 1.
Oligodeoxyribonucleotide Design for Artificial Template IVS2-2A>G Mutation
| Artificial template forward-IVS2-2F (90 bp) |
| 5′ -CAGGTAACTGGTGGTATGGGGCAGTGAGTGCTTCTAGCCTATCCTTGTCGGTGGGTGAATCCCCAGTACGATAGCACCTTCCTGTTTGAC-3′ |
| Artificial template reverse-IVS2-2R (91 bp) |
| 5′ -AGCTAGTTGGAGCCAGGTTACCTGGACTGGGGGCATCAGGCTGCAGAGCTGGGAAGTCGTTGTCAAACAGGAAGGTGCTATCGTACTGGGG-3′ |
The mutation is shown in underline bold italic; the overlapping region is in bold; and short primers are underlined.
PCR Amplification
PCR was performed using GeneAmp 9700 thermal reaction cycler (Applied Biosystems, Foster City, CA). A 20-μl PCR mixture contained 2 μl of extracted DNA (15 to 50 ng/μl), 1.0 μmol/L of each forward and reverse primer, 5′-GCCTGTCCAGTCTTTGAAGC-3′ and 5′-CATTTCGTAGCCAACCATGA-3′ [designed using Primer 321 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi)], 1× FailSafe Premix D containing PCR buffer, dNTPs, and MgCl2 (Epicenter), and 1.0 unit of FastStart DNA polymerase (Roche). Amplification was performed with an initial denaturation at 95°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extension at 72°C for 3 minutes. Unincorporated primers and dNTPs were removed by incubating with 2 units of ExoSAP (USB Corporation) at 37°C for 45 minutes, followed by heat inactivation of enzyme at 80°C for 15 minutes.
Allele-Specific Oligodeoxyribonucleotide Single Nucleotide Extension
Nine extension primers (Integrated DNA Technologies, Inc., Coralville, IA) specific for each mutation were designed to hybridize to either the sense or antisense strand (Table 2). The primers were designed with varying lengths (4 to 5 bp) to adequately separate the multiplexed extended products by capillary electrophoresis. Oligodeoxyribonucleotides longer than 50 bases were purified by high-pressure liquid chromatography. Concentrations were empirically optimized and ranged from 0.1 to 0.4 μmol/L. Two microliters of purified PCR products, 2 μl of the nine pooled allele-specific oligodeoxyribonucleotides extension primers, and 2.5 μl of SNaPshot Multiplex reagent were used to perform a 10-μl SNE assay according to the manufacturer’s protocol (Applied Biosystems). The primers were extended by adding a single fluorescently labeled ddNTP to its 3′-end.22,23,24,25 Fluorescent dyes attached to each ddNTP are as follows: dR6G (green) for A, dTAMRA (yellow, which appears black on the electropherogram) for C, dR110 (blue) for G, and dROX (red) for T. The reactions were cycled with the following conditions: 25 cycles at 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 30 seconds. Samples were treated with one unit of shrimp alkaline phosphatase at 37°C for 45 minutes, followed by heat inactivation of enzyme at 80°C for 15 minutes.
Table 2.
Extension Primer Sequences and Concentrations
| Primer name and variant changes | Primer sequence | Concentration (μM) |
|---|---|---|
| Splice site mutation | 5′-CCTATCCTTGTCGGT-3′ | 0.4 |
| c.253-2A>G | ||
| IVS 2-2A>G | ||
| Exon5 c.404C>T p.S135L | 5′-AGGTCATGTGCTTCCACCCCTGGT-3′ | 0.2 |
| Exon5 c.413C>T* p.T138M | 5′-GGATCTCAGGGACCGACATGAGTGGCAGC-3′ | 0.08 |
| Exon6 c.563A>G p.Q188R | 5′-ATGATGGGCTGTTCTAACCCCCACCCCCACTGCC-3′ | 0.1 |
| Exon7 c.584T>C p.L195P | 5′-CTGCTTTTGCCCCTTGACAGGTATGGGCCAGCAGTTTCC-3′ | 0.36 |
| Exon7 c.626A>G*p.Y209C | 5′-GGCGGCTGTACTCCATTAGCAGGGGCTCTCCATGCTGACTCTTA-3′ | 0.4 |
| Exon7 c.652C>T*†p.L218L | 5′-TTGGCTCTCTCCCACCTTCCTGAGTAGCTCCTGGCGGCTGTACTCCATTA-3′ | 0.16 |
| Exon9 c.285G>T† p.K285N | 5′-GAGTCAGGCTCTGATTCCAGATCTAGCCTCCATCATGAAGAAGCTCTTGACCAA-3′ | 0.12 |
| Exon10 c.940A>G† p.N314D | 5′-CACTGTCTCTCTTCTTTCTGTCAGGGGCTCCCACAGGATCAGAGGCTGGGGCCAACTGG-3′ | 0.4 |
Exon number, nucleotide, and amino acid changes are given with primer sequences and concentrations of nine oligodeoxyribonucleotides.
Primers in italics interrogate the antisense strand.
Primers that are 50-bp long were purified by high-performance liquid chromatography before use.
Two microliters of the fluorescently labeled fragments, 0.25 μl of GeneScan 120 LIZ internal size standard (Applied Biosystems), and 8 μl of HiDi formamide (Applied Biosystems) were resolved by electrophoresis on an automated ABI Prism 3100 Genetic Analyzer using POP-6 (performance-optimized polymer) on a 50-cm array (Applied Biosystems). Samples were electrokinetically injected at 15 kV for 10 seconds and electrophoresed at 15 kV for 1960 seconds at 55°C under filter set E5. Raw data were analyzed with GeneMapper 4.0 (Applied Biosystems) programed for automated calling of both normal and mutant alleles based on fragment size and fluorescence. Allele calls on ABI’s GeneMapper software were automated by calculating the mean and SD of the allele sizes between runs and setting a minimum and maximum size range for each marker. A representative sample of each genotype was confirmed by sequencing (data not shown). For an abnormal sample that did not size within the expected range, bidirectional sequencing was performed.
Results
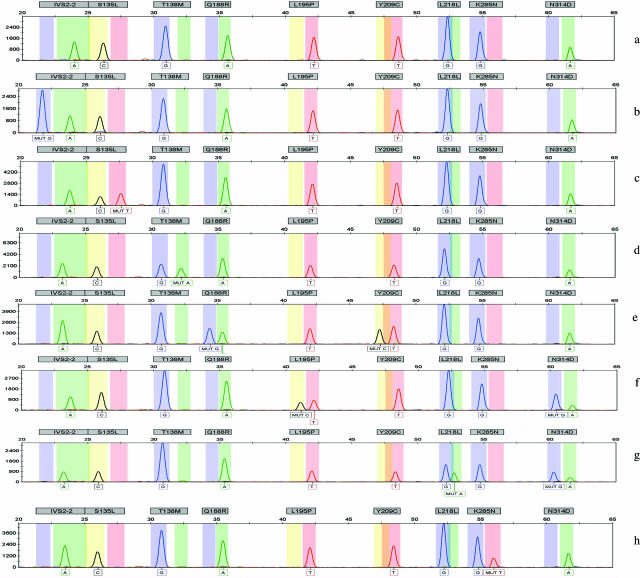
With SNE detection, all nine variants were detected from a single amplicon (Figure 2a2b2c2d2e2f2g2h). The nine single nucleotide extension primers were designed to be different lengths with at least four base differences. The sizes of the oligodeoxyribonucleotides ranged from 15 to 60 bases and were easily distinguished by size during capillary electrophoresis on the ABI 3100 instruments. Each of the nine oligodeoxyribonucleotides independently interrogated the mutation either from the forward or reverse direction depending on the design of the oligodeoxyribonucleotides, incorporating one of the four fluorophore nucleotides into the next nucleotide position.
Figure 2.
Multiplex genotyping of nine common mutations/variants from eight galactosemia samples. A normal wild-type DNA and nine mutations/variants are shown. For each sample, all nine alleles are analyzed and detected as a wild type or a mutant in each size range (bin). Mutations are labeled as “MUT _”. Vertical scales are peak heights measured in relative fluorescent units, and horizontal scales are ranges of oligodeoxyribonucleotides sizes for normal and mutant genotypes detected with each allele. a: Wild-type genotype; b: heterozygous IVS2-2, A>G, artificial template; c: heterozygous p.S135L, C>T; d: heterozygous p.T138M (reverse strand genotyped), G>A mutation; e: compound heterozygous p.Q188R, A>G and p.Y209C, T>C (reverse strand genotyped); f: compound heterozygous p.L195P, T>C and p.N314D, A>G; g: heterozygous p.L218L, G>A (reverse strand genotyped), and p.N314D, A>G; h: heterozygous p.K285N, G>T.
We tested 54 previously genotyped samples (including the artificial template) with 100% concordance with genotypes obtained by the Department of Genetics at the Emory University School of Medicine or ARUP’s allele-specific amplification assay used in the clinical laboratory. The most common mutations were p.Q188R A>G, p.N314D A>G, and p.S135L C>T, similar to other recent studies done in the United States,9 whereas others occurred at lower frequency. Genotypes from samples provided by Emory University were p.L195P/p.N314, heterozygous p.S135L, p.Q188R/p.Y209C, heterozygous p.T138M, heterozygous p.K285N, and heterozygous p.L218L (p.N314D). Genotypes from ARUP clinical samples are summarized in Table 3.
Table 3.
Genotypes of 47 Clinical Samples
| Mutation | Frequency | Percentage |
|---|---|---|
| No mutation/variant detected (–/–) | 13 | 27.6 |
| Q188R/N314D | 10 | 21.3 |
| S135L/N314D | 2 | 4.3 |
| Q188R/Y209C | 1 | 2.1 |
| Q188R/– | 11 | 23.4 |
| N314D/– | 5 | 10.6 |
| N314D/N314D (apparent homozygous) | 3 | 6.4 |
| K285N/– | 1 | 2.1 |
| L218X/– | 1 | 2.1 |
| Total | 47 | ∼100 |
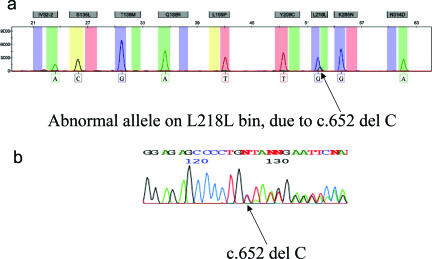
One sample showed one normal allele and one abnormal allele but outside the range validated for the targeted p.L218L mutation (c.652 C>T). On sequencing, this sample was shown to be c.652delC, a novel mutation on exon 7, which is predicted to be a truncated mutation p.L218X (Figure 3).
Figure 3.
a: Sample with nontargeted mutation at locus p.L218L, showing a C base incorporated instead of the expected A base and a shift in the size. b: DNA sequence analysis showing the variant to be p.L218Q due to c.652delC.
From our 47 clinical samples with an abnormal newborn screen, 72% (34 of 47) of the samples had a least one mutation (Table 3), with heterozygotes mutations accounting for 38% (18 of 47 samples) and compound heterozygous mutations for 28% (13 of 47 samples). Twenty-six percent of the samples were D/G compound heterozygotes. Three were apparent homozygous p.N314D mutants, but without testing parents, we cannot exclude hemizygous p.N314D and a gene deletion. In 13 samples (28%), none of the common mutations was detected. Although enzyme activity levels were not available for all samples, seven of the 13 samples with no mutation detected had reduced enzyme activity, suggesting carrier or affected status. These samples would benefit from sequencing the entire GALT gene.
We compared labor and reagents for the SNE assay with the current allele-specific assay performed in our clinical laboratory. By using a long PCR, we reduced the number of reactions from four to one. Reagent costs were reduced approximately 10%. Labor was similar between the two methods because of the amplicon cleanup and the primer extension of SNE. The number of failed reactions requiring repeat testing was reduced from 15 to 5%.
Discussion
When an abnormal screen is identified in a newborn, enzymatic activity confirms the diagnosis for galactosemia. Heterozygous unaffected carriers also may be identified through newborn screening. Combining a mutation panel with enzyme activity identified at least one family-specific mutation or variant in 72% of the cases. Because galactosemia mutations are ethnic-specific,26 we chose mutations that would represent a pan-ethnic population.9,27 Two mutations, p.Q188R and p.K285N, reportedly account for approximately 69 to 88% of galactosemia alleles in Caucasian populations.28 The p.Q188R mutation is considered severe, with no GALT activity in human erythrocytes in the homozygous state.10 p.Q188R also accounts for approximately 50 to 58% of the mutant alleles in Hispanics of Mexican ancestry.29 The IVS2-2A>G is rare but has been described in Hispanic populations and is predicted to be a splice-site variant.9 The p.K285N mutation is also severe but has a frequency of approximately 26% in mutant alleles in Eastern Europeans.10,30 The p.S135L is the most common mutation in African Americans and accounts for 50% of the mutations observed in that population. This mutation is usually associated with a milder clinical phenotype than p.Q188R, which is the second most common mutation in the Black population.27 Other mutations included in the panel had lower frequencies but still represented over 1% of the galactosemia alleles. These were p.L195P (2.6%),2,10,17,31,32 p.Y209C (1.3%),28 and p.T138M (2%).9,28,33 The p.T138M has been described in a galactosemia patient with a milder phenotype.33
By including the LA (which increases activity) and Duarte (which mildly reduces activity) variants, this panel also clarified intermediate activities by detecting a combination of known mutations and genetic variants that modify enzyme levels. Compound heterozygous individuals for Duarte variant and one classic galactosemia allele (D/G) will have approximately 25 to 30% of normal GALT enzyme activity, creating a partial reduction in GALT enzyme activity. In most D/G individuals, the condition is benign, but some individuals can have symptoms resembling classic galactosemia. One possible explanation for phenotypic variability among D/G individuals is modification of the Duarte variant by additional variants, for example, the E203K variant that may stabilize the protein.34
Advantages of SNE include straightforward design and easy optimization. It is also capable of multiplexing and simultaneously detecting multiple mutations or variants in a single reaction. We designed a PCR assay that amplified 2115 bp encompassing all of the seven mutations and two variants. The SNE assay was then used to detect all of the variants present within each sample simultaneously. This SNE assay has the advantage of being simple and accurate in distinguishing simple and compound heterozygous and homozygous genotypes. Automatic base calling on GeneMapper software (Applied Biosystems) based on fragment size and fluorescence on both normal and mutant alleles further reduces labor. Samples with an abnormal allele but with a different extended base or outside the defined range should be sequenced to confirm a nontargeted mutation. The main advantage of this method is a reduced failure rate, thus improving patient care by more rapid turnaround times. This assay with these combinations should rapidly and economically detect and specify the mutation involved in over 80% of individuals with some form of galactosemia. Used as a confirmatory screening method, it can supplement the enzymatic assay for classic galactosemia.
Acknowledgments
We gratefully acknowledge Friederike Gedge for her help with GeneMapper 4.0, Jacquelyn McCowen-Rose for her help in scanning photos, Dr. Kasinathan Muralidhara of Emory University School of Medicine, Department of Genetics, and the ARUP Laboratories for supplying GALT samples.
References
- Leslie ND. Insights into the pathogenesis of galactosemia. Annu Rev Nutr. 2003;23:59–80. doi: 10.1146/annurev.nutr.23.011702.073135. [DOI] [PubMed] [Google Scholar]
- Tyfield L, Reichardt J, Fridovich-Keil J, Croke DT, Elsas 2nd LJ, Strobl W, Kozak L, Coskun T, Novelli G, Okano Y, Zekanowski C, Shin Y, Boleda MD. Classical galactosemia and mutations at the galactose-1-phosphate uridyl transferase (GALT) gene. Hum Mutat. 1999;13:417–430. doi: 10.1002/(SICI)1098-1004(1999)13:6<417::AID-HUMU1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Beutler E. Galactosemia: screening and diagnosis. Clin Biochem. 1991;24:293–300. doi: 10.1016/0009-9120(91)80003-l. [DOI] [PubMed] [Google Scholar]
- Beutler E, Baluda MC, Sturgeon P, Day R. A new genetic abnormality resulting in galactose-1-phosphate uridyltransferase deficiency. Lancet. 1965;1:353–354. doi: 10.1016/s0140-6736(65)91782-4. [DOI] [PubMed] [Google Scholar]
- Beutler E, Baluda MC. Improved method for measuring galactose-I-phosphate uridyl transferase activity of erythrocytes. Clin Chim Acta. 1966;13:369–379. doi: 10.1016/0009-8981(66)90217-8. [DOI] [PubMed] [Google Scholar]
- Mellman WJ, Tedesco TA. An improved assay of erythrocyte and leukocyte galactose-1-phosphate uridyl transferase: stabilization of the enzyme by a thiol protective reagent. J Lab Clin Med. 1965;66:980–986. [PubMed] [Google Scholar]
- Stambolian D. Galactose and cataract. Surv Ophthalmol. 1988;32:333–349. doi: 10.1016/0039-6257(88)90095-1. [DOI] [PubMed] [Google Scholar]
- Shurin SB. Escherichia coli septicemia in neonates with galactosemia. N Engl J Med. 1977;297:1403–1404. doi: 10.1056/NEJM197712222972516. [DOI] [PubMed] [Google Scholar]
- Yang YP, Corley N, Garcia-Heras J. Molecular analysis in newborns from Texas affected with galactosemia. Hum Mutat. 2002;19:82–83. doi: 10.1002/humu.9005. [DOI] [PubMed] [Google Scholar]
- Greber-Platzer S, Guldberg P, Scheibenreiter S, Item C, Schuller E, Patel N, Strobl W. Molecular heterogeneity of classical and Duarte galactosemia: mutation analysis by denaturing gradient gel electrophoresis. Hum Mutat. 1997;10:49–57. doi: 10.1002/(SICI)1098-1004(1997)10:1<49::AID-HUMU7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Elsas LJ, Dembure PP, Langley S, Paulk EM, Hjelm LN, Fridovich-Keil J. A common mutation associated with the Duarte galactosemia allele. Am J Hum Genet. 1994;54:1030–1036. [PMC free article] [PubMed] [Google Scholar]
- Lin HC, Kirby LT, Ng WG, Reichardt JK. On the molecular nature of the Duarte variant of galactose-1-phosphate uridyl transferase (GALT). Hum Genet. 1994;93:167–169. doi: 10.1007/BF00210604. [DOI] [PubMed] [Google Scholar]
- Elsas LJ, Lai K, Saunders CJ, Langley SD. Functional analysis of the human galactose-1-phosphate uridyltransferase promoter in Duarte and LA variant galactosemia. Mol Genet Metab. 2001;72:297–305. doi: 10.1006/mgme.2001.3157. [DOI] [PubMed] [Google Scholar]
- Kozák L, Francova H, Pijackova A, Macku J, Stastna S, Peskovova K, Martincova O, Krijt J. Presence of a deletion in the 5′ upstream region of the GALT gene in Duarte (D2) alleles. J Med Genet. 1999;36:576–578. [PMC free article] [PubMed] [Google Scholar]
- Guthrie R. The origin of newborn screening. Screening. 1992;1:5–15. doi: 10.1016/0925-6164(92)90025-z. [DOI] [PubMed] [Google Scholar]
- Beutler E, Baluda MC, Halasz A. Biochemical properties of human red cell galactose-1-phosphate uridyl transferase (UDP glucose: alpha-D-galactose-1-phosphate uridyltransferase E.C. 2.7.7.12) from normal and mutant subjects. J Lab Clin Med. 1966;67:947–954. [PubMed] [Google Scholar]
- Item C, Hagerty BP, Muhl A, Greber-Platzer S, Stockler-Ipsiroglu S, Strobl W. Mutations at the galactose-1-p-uridyltransferase gene in infants with a positive galactosemia newborn screening test. Pediatr Res. 2002;51:511–516. doi: 10.1203/00006450-200204000-00018. [DOI] [PubMed] [Google Scholar]
- Angelaccio S, Bonaccorsi di Patti MC. Site-directed mutagenesis by the megaprimer PCR method: variations on a theme for simultaneous introduction of multiple mutations. Anal Biochem. 2002;306:346–349. doi: 10.1006/abio.2002.5689. [DOI] [PubMed] [Google Scholar]
- Burke E, Barik S. Megaprimer PCR: application in mutagenesis and gene fusion. Methods Mol Biol. 2003;226:525–532. doi: 10.1385/1-59259-384-4:525. [DOI] [PubMed] [Google Scholar]
- Ling MM, Robinson BH. Approaches to DNA mutagenesis: an overview. Anal Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yu YX, Liu XX, Wei L. [SNaPshot technique for detection of single-nucleotide polymorphisms (SNPs) in HBV polymerase gene region of HBV gene]. Chinese. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2005;19:162–164. [PubMed] [Google Scholar]
- Grignani P, Peloso G, Achilli A, Turchi C, Tagliabracci A, Alu M, Beduschi G, Ricci U, Giunti L, Robino C, Gino S, Previdere C. Subtyping mtDNA haplogroup H by SNaPshot minisequencing and its application in forensic individual identification. Int J Legal Med. 2006;120:151–156. doi: 10.1007/s00414-005-0059-5. [DOI] [PubMed] [Google Scholar]
- Quintáns B, Alvarez-Iglesias V, Salas A, Phillips C, Lareu MV, Carracedo A. Typing of mitochondrial DNA coding region SNPs of forensic and anthropological interest using SNaPshot minisequencing. Forensic Sci Int. 2004;140:251–257. doi: 10.1016/j.forsciint.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Turner D, Choudhury F, Reynard M, Railton D, Navarrete C. Typing of multiple single nucleotide polymorphisms in cytokine and receptor genes using SNaPshot. Hum Immunol. 2002;63:508–513. doi: 10.1016/s0198-8859(02)00392-0. [DOI] [PubMed] [Google Scholar]
- Suzuki M, West C, Beutler E. Large-scale molecular screening for galactosemia alleles in a pan-ethnic population. Hum Genet. 2001;109:210–215. doi: 10.1007/s004390100552. [DOI] [PubMed] [Google Scholar]
- Lai K, Langley SD, Singh RH, Dembure PP, Hjelm LN, Elsas LJ., 2nd A prevalent mutation for galactosemia among black Americans. J Pediatr. 1996;128:89–95. doi: 10.1016/s0022-3476(96)70432-8. [DOI] [PubMed] [Google Scholar]
- Elsas LJ, 2nd, Lai K. The molecular biology of galactosemia. Genet Med. 1998;1:40–48. doi: 10.1097/00125817-199811000-00009. [DOI] [PubMed] [Google Scholar]
- Ng WG, Xu YK, Kaufman FR, Donnell GN, Wolff J, Allen RJ, Koritala S, Reichardt JK. Biochemical and molecular studies of 132 patients with galactosemia. Hum Genet. 1994;94:359–363. doi: 10.1007/BF00201593. [DOI] [PubMed] [Google Scholar]
- Kozak L, Francova H, Fajkusova L, Pijackova A, Macku J, Stastna S, Peskovova K, Martincova O, Krijt J, Bzduch V. Mutation analysis of the GALT gene in Czech and Slovak galactosemia populations: identification of six novel mutations, including a stop codon mutation (X380R). Hum Mutat. 2000;15:206. doi: 10.1002/(SICI)1098-1004(200002)15:2<206::AID-HUMU13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lai K, Elsas LJ. Structure-function analyses of a common mutation in blacks with transferase-deficiency galactosemia. Mol Genet Metab. 2001;74:264–272. doi: 10.1006/mgme.2001.3230. [DOI] [PubMed] [Google Scholar]
- Bosch AM, Ijlst L, Oostheim W, Mulders J, Bakker HD, Wijburg FA, Wanders RJ, Waterham HR. Identification of novel mutations in classical galactosemia. Hum Mutat. 2005;25:502. doi: 10.1002/humu.9330. [DOI] [PubMed] [Google Scholar]
- Shin YS, Gathof BS, Podskarbi T, Sommer M, Giugliani R, Gresser U. Three missense mutations in the galactose-1-phosphate uridyltransferase gene of three families with mild galactosaemia. Eur J Pediatr. 1996;155:393–397. doi: 10.1007/BF01955270. [DOI] [PubMed] [Google Scholar]
- Elsas LJ, Langley S, Steele E, Evinger J, Fridovich-Keil JL, Brown A, Singh R, Fernhoff P, Hjelm LN, Dembure PP. Galactosemia: a strategy to identify new biochemical phenotypes and molecular genotypes. Am J Hum Genet. 1995;56:630–639. [PMC free article] [PubMed] [Google Scholar]