Abstract
The goal of this study was to compare how accumulation of chromosomal aberrations in human papillomavirus (HPV)-infected cells correlates with the severity of cervical dysplastic lesions. We assessed the frequency of genomic alterations for 35 different loci in a pilot biopsy study and selected two loci (3q26 and 8q24) with the highest frequency of copy number gains found in high-grade dysplasia and cancer. These probes were labeled with gold and red fluorophores and combined with HPV biotin-labeled probes for subsequent detection using a tyramide signal amplification system with a green fluorophore. Cells that were both HPV positive and chromosomally abnormal were designated as “double-positive cells.” Cervical cytology specimens from 235 patients were used for this blinded study. The average number of double-positive cells increased from two cells in patients with a cytological interpretation of atypical squamous cells of undetermined significance to 22 cells in low-grade squamous intraepithelial lesion and 99 cells in high-grade squamous intraepithelial lesion, reflecting an accumulation of chromosomal abnormality with disease progression. Using a cutoff of four or more double-positive cells as the criterion for the presence of a cervical intraepithelial neoplasia 2 or 3 lesion, we demonstrated that low-grade squamous intraepithelial lesion and high-grade squamous intraepithelial lesion cytology specimens with underlying cervical intraepithelial neoplasia 2/3 histology showed positive test results in more than 80% of cases. Correlation of 3q26 and 8q24 aneusomy with concurrent HPV infection may thus serve as a biomarker of genetic instability in HPV-infected cells.
In the last decade, the etiological role of human papillomavirus (HPV) infection in the development of cervical dysplasia and cancer has been well established.1,2,3,4 The frequency of HPV infection in women with a diagnosis of atypical squamous cells of undetermined significance (ASCUS), low-grade squamous intraepithelial lesion (LSIL), or high-grade squamous intraepithelial lesion (HSIL) is approximately 50, 80, and 95%, respectively.5,6,7 Still, only a fraction of HPV-infected women will develop high-grade lesions and cervical cancer.8
HPV infections compromise normal cellular proliferation through degradation of the tumor suppressor proteins p53 and pRB by viral proteins E6 and E7, respectively. In addition, HPV infection has been shown to induce abnormal centrosome duplication early in the infection process.9,10,11,12 HPV infection of replicating immature cells prevents epithelial maturation and differentiation, leading to continued replication and accumulation of genetic abnormalities.2,13,14
Chromosomal instability at a numerical or structural level is a hallmark of malignant tumors.9 Deletion, duplication, and amplification of various genomic regions have been demonstrated in cervical cancer by comparative genomic hybridization and fluorescence in situ hybridization (FISH) methods.15,16,17,18 In an internal study, we assessed biopsy specimens showing high-grade dysplasia and cancer with FISH probes to 35 unique loci and identified 2 loci, the 3q26 region (comprising H-TERC gene) and the 8q24 region (comprising c-MYC gene), which showed highest frequency of copy number gains in high-grade dysplasia and cancer. Because these loci are frequently altered in cervical cancer tumorigenesis,16,17,19,20 we hypothesized that they might be useful markers for the detection of cervical dysplasia and carcinoma.
To explore this further, we created a fluorescence in situ hybridization assay that allows for the simultaneous detection of HPV and numerical alterations on chromosome regions 3q26 and 8q24 in cervical cytology specimens and evaluated it on 235 cervical cytology specimens from women with a concurrent biopsy.
Materials and Methods
Cervical Specimens
Residual cervical specimens were obtained from the University of Washington, Mayo Clinic, and Cleveland Clinic in PreservCyt solution (Cytyc, Inc., Marlborough, MA). The specimens had been archived from 3 months to 3 years before being used in this study. ThinPrep slides were prepared using the ThinPrep 2000 Processor (Cytyc) following the manufacturer’s instructions. Slides were dried overnight at room temperature and then stored at −20°C until hybridized. Specimens with an abnormal cytological diagnosis (ASCUS, LSIL, and HSIL) were included in the study if they had a corresponding biopsy diagnosis within 4 months of the cytological diagnosis. Cytology specimens with a cytological diagnosis of “normal” were also included, but not all of these patient specimens had a corresponding biopsy according to the standard practice of care. The distribution of specimens based on the cytology and histology classification was as follows: 15 cases with normal cytology and no biopsy available, 11 cases with normal cytology and normal biopsy, 14 cases with ASCUS cytology and cervical intraepithelial neoplasia 2/3 (CIN2/3) biopsy, 42 cases with LSIL cytology and normal biopsy, 68 cases with LSIL cytology and CIN1 biopsy, 37 cases with LSIL cytology and CIN2/3 biopsy, and 48 cases with HSIL cytology and CIN2/3 biopsy. Evaluation of slides after FISH assay was performed without knowledge of patient information.
Probe Set Formulation
The following HPV DNAs were included in the probe set: HPV16, HPV18, HPV30, HPV45, HPV51, and HPV58. The selection of HPV types for use in the probe set was determined by high-risk classification or high homology to high-risk HPV types and commercial availability. HPV16 and HPV18 were both obtained from American Type Culture Collection (Manassas, VA). HPV30 and HPV45 were obtained from Dr. Ethel-Michele de Villiers from DKFZ (Heidelberg, Germany). HPV51 and HPV58 were obtained from Dr. Klara Abravaya (Abbott Molecular, Inc., Des Plaines, IL). HPV51 was provided to Dr. Klara Abravaya by Dr. Sara Gusik (Columbia University, NY). HPV58 was provided to Dr. Klara Abravaya by Dr. Motoyasu Sugase (Nagano Red Cross Hospital, Nagano, Japan).
Pilot experiments using clinical specimens with known HPV types confirmed the detection of the following HPV types: HPV16, HPV18, HPV26, HPV31, HPV33, HPV35, HPV39, HPV45, HPV52, HPV53, HPV56, HPV59, HPV66, and HPV82. No specimens were available with HPV51. However, specimens infected with HPV82, a type that shares about 80% homology with HPV51, were successfully detected. Based on a BLAST2 sequence analysis, the types that were detected in this study had at least 50% homology to the HPV DNAs used in the probe set.
For convenience, all HPV DNA included in our probe cocktail were recloned into the pBluescript SKII (−) plasmid (Stratagene, La Jolla, CA). HPV constructs that included both insert and the vector were individually labeled with biotin by means of nick-translation using a kit from Invitrogen (Carlsbad, CA) so that the median size of the final product was about 100 to 400 bp. Biotin-labeled pBluescript was tested in hybridization experiments on various cell lines and clinical specimens as a negative control probe to ensure specificity of the HPV probe signal. In addition, each HPV probe was tested individually to confirm that each probe was detecting the expected HPV type. The percentage of biotin incorporation was determined for each biotinylated probe using the FluoReporter Biotin quantitation assay kit (Molecular Probes, Eugene, OR) and was approximately 1 to 3%.
The locus-specific probes 3q26 and 8q24 were BAC contig probes and were manufactured by Vysis/Abbott (Des Plaines, IL) using a standard labeling procedure. The chromosome probe 3q26 (TERC) was labeled with the SpectrumGold fluorophore, and the 8q24 probe (MYC) was labeled with the SpectrumRed fluorophore. The six biotinylated HPV probes and the two locus-specific chromosome probes (TERC and MYC) were then combined into a single probe mix containing LSI buffer (Vysis/Abbott), blocking DNA, and modified salt concentration to ensure HPV probes hybridization to most of the high-risk HPV DNA. The final composition of the hybridization cocktail was as follows: LSI hybridization buffer from Vysis/Abbott, 20× standard saline citrate (SSC), Cot-1 DNA, human placental DNA, and biotinylated HPV DNAs (HPV types 16, 18, 30, 45, 51, and 58). Each reaction contained 2 μg of human placental DNA, 1 μg of Cot-1 DNA, 90 ng of SpectrumRed MYC probe, 100 ng of SpectrumGold TERC probe, and 25 ng of each biotinylated HPV DNA (150 ng total HPV DNA) in 4× SSC (final concentration).
Sample Pretreatment and Hybridization
ThinPrep slides were soaked in 2× SSC at 73°C for 2 minutes and then incubated in pepsin (0.5 mg/ml in 10 mmol/L HCl) at 37°C for 10 minutes. The slides were then soaked in 1× PBS at room temperature for 5 minutes, fixed in 1% neutral-buffered formalin at room temperature for 5 minutes, and soaked in 1× PBS for 5 minutes. Slides were dehydrated in an ethanol series of 70, 85, and 100% for 1 minute in each solution and air-dried. The probe mixture was then applied, and slides were coverslipped and sealed with rubber cement. The slides with probe mix were co-denatured at 72°C for 2 minutes and then hybridized at 37°C for 16 to 18 hours on a HyBrite or ThermoBrite (Vysis/Abbott). After hybridization, slides were washed in 2× SSC/0.3% Nonidet P-40 for 2 minutes at 48°C and then in 2× SSC/0.1% Nonidet P-40 for 1 minute at room temperature.
Tyramide Signal Amplification Assay for HPV Probe Development
Detection of the biotinylated HPV probes was performed using the Alexa Fluor 488 TSA (tyramide signal amplification) kit number 22 (Molecular Probes) following the manufacturer’s directions. Endogenous peroxidase activity was blocked by incubation in 3% H2O2 for 30 minutes at room temperature. Slides were washed in 1× PBS for 5 minutes at room temperature. Slides were then incubated with 1% blocking reagent in PBS at 37°C for 25 minutes followed by streptavidin-horseradish peroxidase (1:100 dilution in blocking reagent) at 37°C for 25 minutes (both incubations were performed in a humidified chamber). After washing the slides three times in 1× PBS for 5 minutes at 37°C, the biotin-labeled HPV probe streptavidin-horseradish peroxidase complex was visualized by incubation with Alexa Fluor 488-labeled tyramide (1:100 dilution in amplification buffer, containing 0.0015% H2O2) for 10 minutes at room temperature (in a humidified chamber). The slides were then washed three times in 1× PBS for 5 minutes at 37°C; the nuclear counterstain 4,6-diamidino-2-phenylindole (DAPI) was applied, and slides were coverslipped.
Examination of Clinical Specimens after FISH
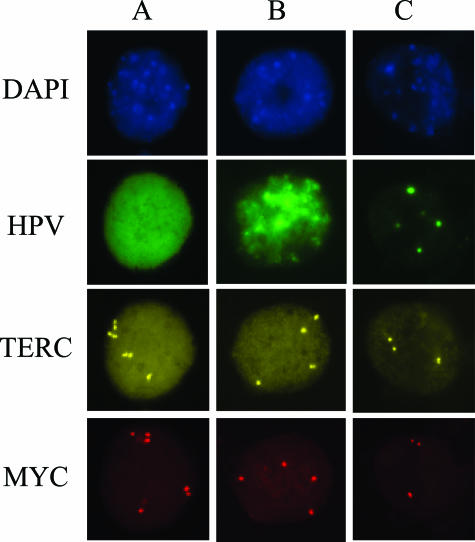
After tyramide development, slides were analyzed under a fluorescence microscope using 40× magnification and single bandpass filter sets. Cell nuclei were visualized with a DAPI filter (Figure 1, DAPI). HPV-positive cells were visualized using a green filter (Figure 1, HPV). HPV staining was observed as diffuse, mixed (a combination of both diffuse and punctate), or punctate (Figure 1, HPV-A, HPV-B, and HPV-C, respectively). Diffuse staining, suggestive of episomal HPV state, is defined as complete green staining of the nucleus (Figure 1, HPV-A). Conversely, punctate staining, suggestive of integrated HPV state, is limited to one or several individual spots of green staining (Figure 1, HPV-C). True HPV staining was localized to the nucleus as confirmed by DAPI staining and co-localization with the locus-specific probes. The 8q24 probe was visualized with a red filter set (Figure 1, MYC), and the 3q26 probe was visualized with a gold filter set (Figure 1, TERC). All filter sets are from Abbott Molecular Inc.
Figure 1.
A representative image of HPV staining and chromosome probes 3q26 (TERC) and 8q24 (MYC) signals observed in cervical epithelial cells after FISH assay. Each individual signal for the TERC and MYC probes constitutes one copy of the respective gene. The images consist of DAPI staining of cell nuclei, HPV staining (green) of infected cells, chromosome probe 3q26 (TERC) visualized using a gold filter, and chromosome probe 8q24 (MYC) visualized using a red filter. Column A represents a cell episomally infected with HPV, column B represents a cell with a mixed HPV infection, and column C represents an integrated HPV infection.
In most cases, the entire surface area of each slide was analyzed. However, in cases involving a large number of HPV-positive cells per slide, only the first 100 HPV-positive cells were analyzed. In these cases the estimated number of HPV-positive cells on the entire slide was extrapolated from the percentage of surface area enumerated after enumerating 100 HPV-positive cells. Screening of slides was conducted by beginning at one corner of the slide and advancing from one field of view to the next in an orderly manner observing and documenting all nuclear staining through the green filter. The number of gold probe (3q26) fluorescent signals and red probe (8q24) fluorescent signals was recorded for each HPV-positive cell. Cells that were both HPV positive and chromosomally abnormal were designated as “double-positive cells.”
Data Analysis
Enumeration results were analyzed using JMP statistical software, version 5 (SAS Institute Inc., Cary, NC). P values <0.05 were considered statistically significant.
Results
Evaluation of 8q24 and 3q26 Probe Copy Number Changes in HPV-Infected Cells of Cytological Specimens
To evaluate chromosomal changes, HPV-positive cells identified on the slide were analyzed for 3q26 and 8q24 copy number. An image representing various types of HPV staining and chromosomal abnormalities is shown in Figure 1. The images were obtained from samples that were classified as having high-grade histology (CIN2 and CIN3). A cell was considered chromosomally abnormal if either the 3q26 or the 8q24 probe showed three or more signals per cell (ie, gains of these loci). For enumeration, each visible signal corresponded to one copy of the gene being detected. All three nuclei depicted in Figure 1 (columns A, B, and C) were considered to be chromosomally abnormal. The nucleus in Figure 1, column A, had seven copies of the TERC probe and six copies of the MYC probe. The chromosomal counts for the nucleus found in Figure 1, column B, had four TERC signals and four MYC signals. The nucleus in Figure 1, column C, had three TERC and three MYC signals.
The average copy number for each chromosome probe in HPV-infected cells was calculated for each cytological category (Table 1). There was an increase in copy number for both chromosome markers among patients with increasing severity of cytological diagnosis. The TERC probe revealed an increase from 2.17 copies per cell for cytologically normal patients to 2.63 copies per cell for patients diagnosed with high-grade dysplasia. Similarly, the MYC probe exhibited an increase from 2.17 to 2.61 copies per cell from cytologically normal patients to patients with high-grade dysplasia, respectively. The majority of chromosomally abnormal cells showed a low-level copy number gain, ie, three to four signals, in both 3q26 and 8q24 chromosome regions in HPV-infected cells. Occasional cells were found to display five to eight copies of each probe. Approximately 10% of all HPV-infected cells carrying chromosomal aberrations were found to have amplification in only one of the two chromosomal loci, either 3q26 probe or 8q24 probe.
Table 1.
Average Number of Copies per Cell for Chromosome Probe Markers 3q26 (TERC) and 8q24 (MYC) in a Population of HPV-Infected Cervical Epithelial Cells for Different Cytological Categories
| Cytological category | Mean 3q26 probe (SEM) | Mean 8q24 probe (SEM) |
|---|---|---|
| Normal | 2.17 (0.17) | 2.17 (0.17) |
| ASCUS | 2.47 (0.06) | 2.49 (0.05) |
| LSIL | 2.54 (0.01) | 2.59 (0.01) |
| HSIL | 2.63 (0.01) | 2.61 (0.01) |
Relationships between HPV-Positive Cells and Chromosomal Alterations
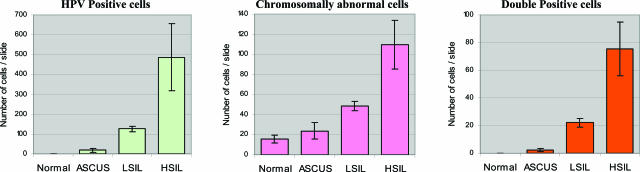
The quantification results for HPV-positive, chromosomally abnormal, and double-positive cells in cervical cytology specimens grouped by cytology diagnosis are shown in Figure 2. This analysis reveals that the average number of HPV-positive cells increases accordingly with escalating levels of dysplasia. The patients classified as cytologically normal did not have any HPV-positive cells, whereas those classified as LSIL and HSIL had, on average, 125 and 487 HPV-positive cells, respectively. In addition, the samples with an ASCUS diagnosis had an average of 19 HPV-positive cells. Further extrapolation of the data reveals that in thin-layer Pap smear specimens, approximately 1 in 1250 cells is infected with HPV in the ASCUS group, 1 in 200 cells in the LSIL group, and 1 in 50 cells in the HSIL group (based on approximately 23,000 cells/slide). The difference in the quantity of HPV-infected cells in each of these categories was statistically significant (P < 0.05; Table 2).
Figure 2.
Average quantity of HPV-positive cells, chromosomally abnormal cells, and double-positive cells per cytological category.
Table 2.
Statistical Significance Determined for Different Cytological and Histological Groups
|
P value
|
|||
|---|---|---|---|
| HPV- positive cells | Chromosomally abnormal cells | Double- positive cells | |
| Cytology groups | |||
| NIL versus ASCUS | 0.0072 | 0.382 | 0.003 |
| NIL versus LSIL | 0.005 | 0.009 | 0.0039 |
| NIL versus HSIL | 0.037 | 0.019 | 0.0061 |
| ASCUS versus LSIL | 0.029 | 0.047 | 0.057 |
| LSIL versus HSIL | 0.0003 | 0.0004 | <0.0001 |
| Histology groups | |||
| CIN1 versus CIN2 | 0.072 | 0.092 | 0.028 |
| CIN1 versus CIN3 | 0.014 | 0.146 | 0.0272 |
The number of chromosomally abnormal cells per slide (regardless of HPV status) was also determined for a randomly selected subset of all of the cases from each of the different cytological categories. The results of this enumeration revealed that, on average, 15 cells with chromosomal gains (in at least one chromosomal locus) were found in patients with normal cytology, representing 0.07% of cells per slide. For the other cytological categories, an average of 23 cells with chromosome gains for either chromosomal locus were found in the ASCUS category, 49 cells in the LSIL category, and an average of 101 chromosomally abnormal cells in the HSIL category, representing 0.1, 0.2, and 0.44% of total population of cells per slide, respectively.
For each HPV-positive stained cell on a slide, we recorded the chromosome copy number for TERC and MYC probes. Cells that were both HPV positive and chromosomally abnormal were designated as “double-positive cells.” The results show that the average number of double-positive cells per slide increased with cytological diagnosis: 2 in ASCUS, 22 in LSIL, and 99 in HSIL samples (for comparison of statistical significance, see Table 2). No double-positive cells were found in the group of cytologically normal samples.
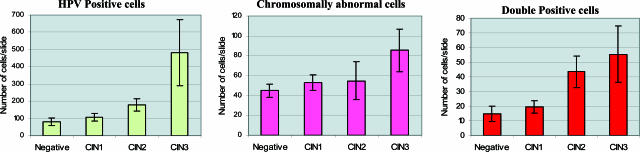
Figure 3 presents the results according to histological classification. The samples categorized with CIN1 lesions had an average of 108 HPV-positive cells per slide. A marked increase in the number of HPV-positive cells was observed when compared with the samples with CIN2 lesions, which had an average of 179 HPV-positive cells per slide. The difference observed between the samples diagnosed as CIN2 and CIN3 was even more drastic, with 179 and 482 HPV-positive cells per slide, respectively.
The average number of chromosomally abnormal cells found on slides from the CIN1 and CIN2 categories was similar. The samples with a CIN1 diagnosis had an average of 53 aneusomic cells per slide and the CIN2 slides had 55. The group of CIN3 samples, however, revealed a significant increase to 85 chromosomally abnormal cells per slide. The differences between these diagnostic categories were not statistically significant (Table 2).
The number of double-positive cells increased substantially between CIN1 and CIN2 categories, from 18 double-positive cells to 43 double-positive cells (P = 0.028). The increase in the number of double-positive cells was less pronounced between CIN2 and CIN3 categories (43 to 55 cells, respectively; P = 0.569).
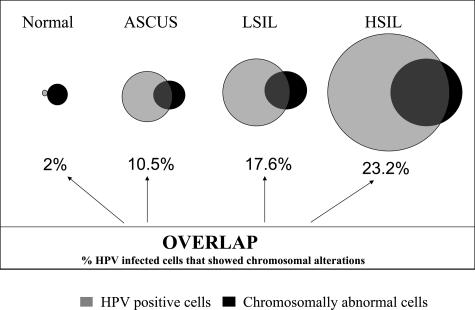
The relationships between HPV infection, chromosomal gains, and cytological diagnosis are depicted in Figure 4. Not all chromosomally abnormal cells were found to be positive by HPV, and not all HPV-positive cells show signs of genomic instability. It was determined that 10.5% of HPV-infected cells in the ASCUS category carried chromosomal gains, whereas 17.6% had chromosomal gains in LSIL samples, and 23.2%, in HSIL samples. Overall, the advancement of the disease correlated well with the accumulation of HPV-infected cells and the increased genomic instability in HPV-infected cells.
Figure 4.
Relationships between HPV-infected and chromosomally abnormal cervical epithelial cells depicted in a Venn diagram. The numbers beneath the diagram circles reflect the percentage of HPV-infected cells that demonstrated chromosome copy gains for either one or both chromosome markers (3q26 and 8q24 probes).
To futher investigate the relationships between HPV-positive cells and chromosomal abnormality, we analyzed the data based on the results that were available for both cytological and histological evaluations (Table 3). Samples that were found to be cytologically normal and histologically negative (normal/negative) or did not have a biopsy performed (normal/NA) did not have any double-positive cells nor did they have any HPV-positive cells. The samples with ASCUS cytology and CIN2 histology (ASCUS/CIN2) had an average of 28 HPV-positive cells per slide and three double-positive cells per slide; the ASCUS/CIN3 samples averaged 12 HPV-positive and two double-positive cells per slide.
Table 3.
Comparison of the Quantity of HPV-Positive and Double-Positive Cells Categorized According to Their Cytological and Histological Findings
| Cytological category (n) | Histology category | Average number HPV-positive cells per slide | Average number double-positive cells per slide |
|---|---|---|---|
| Normal (15) | Biopsy not performed | 0 | 0 |
| Normal (11) | Negative | 0 | 0 |
| ASCUS (6) | CIN2 | 28 | 3 |
| ASCUS (8) | CIN3 | 12 | 2 |
| LSIL (42) | Negative | 103 | 19 |
| LSIL (68) | CIN1 | 108 | 19 |
| LSIL (32) | CIN2 | 190 | 33 |
| LSIL (5) | CIN3 | 148 | 15 |
| HSIL (19) | CIN2 | 208 | 73 |
| HSIL (29) | CIN3 | 669 | 77 |
The specimens diagnosed cytologically as LSIL encompassed negative, CIN1, CIN2, and CIN3 histological classifications. Interestingly, the subgroup of patients with LSIL diagnosis and negative histological findings demonstrated results very similar to the LSIL cytology group with a CIN1 histological subgroup. In the LSIL/negative and LSIL/CIN1 groups, the average number of HPV-positive cells per slide was 103 and 108 cells, respectively. The average number of double-positive cells was 19 cells in each group. In the subgroup of LSIL/CIN2, a marked increase in HPV-positive cells was observed along with the increase of double-positive cells (190 HPV-positive cells and 33 double-positive cells; Table 3). Although the LSIL/CIN3 category shows a decrease in the number of HPV-positive cells and double-positive cells per slide (148 and 15, respectively), this subgroup was poorly represented in our study because it was limited to only five specimens. Therefore, quantitative findings for this subgroup should be interpreted with care.
The HSIL cytological category comprised CIN2 and CIN3 histological diagnoses. The HSIL/CIN2 group had an average of 208 HPV-positive cells per slide, which was nearly identical to the LSIL/CIN2 group (190 HPV-positive cells per slide). However, the number of double-positive cells for the HSIL/CIN2 group (73) exhibited more than a twofold difference when compared with the LSIL/CIN2 group (33). A drastic increase in the accumulation of HPV-infected cells was observed in the HSIL subgroup with CIN3 histology compared with the HSIL/CIN2 group: 669 HPV-positive cells versus 208 HPV-positive cells, respectively. The data show that a further increase in the total quantity of HPV-infected cells (up to threefold) did not correspond to a substantial accumulation of chromosomal aberrations: the total number of double-positive cells in HSIL/CIN3 group was almost identical to what was found in HSIL/CIN2.
Clinical Correlations between Double-Positive Cells and Disease Categories
The utility of double-positive cell evaluation to detect lesions with a CIN2 diagnosis or higher was investigated. Various cutoff values for double-positive cells were evaluated to determine the number of double-positive cells that would properly segregate CIN2/3 lesions from normal and CIN1 lesions. A sample was considered positive for the test if the number of double-positive cells was equal to or higher than the cutoff value. For the data presented in Table 4, a cutoff value of four double-positive cells was used. None of the samples with normal cytology and negative histology was positive for HPV in this study. Using a cutoff of four double-positive cells for the standard assay, we determined that in CIN2 and CIN3 subgroups of ASCUS, the assay positivity ranged from 25 to 33%. A surprisingly high rate of positive results was observed in LSIL patients with negative and mild histology findings. The test was positive in 45% of the LSIL/negative subgroup and in 56% of patients from the LSIL/CIN1 subgroup. The rate of assay positivity was 81% in the LSIL/CIN2 subgroup and 80% in the LSIL/CIN3 subgroup. In the latter group, the number of cases evaluated was small (five cases) and should be considered with caution. The HSIL/CIN2 and HSIL/CIN3 subgroups were positive in 78 and 86% of cases, respectively. Some of these samples were retested with a more sensitive version of the assay using a double-tyramide amplification step (data not shown). Based on the cutoff of four double-positive cells, this converted a few of the cases from negative to positive and brought the resulting assay positivity rate in HSIL/CIN3 to 93%. It is important to note that two of the HSIL/CIN3 patients still remained negative after the re-test and displayed a high degree of chromosomal abnormality with many cells carrying five to seven copies for each chromosome probe.
Table 4.
Cervical Test Positivity Rate For Different Clinical Groups
| Cytology-histology | Test-positive cases [% (positive/total)] |
|---|---|
| ASCUS-CIN2 | 33 (2/6) |
| ASCUS-CIN3 | 25 (2/8) |
| LSIL-negative | 45 (19/42) |
| LSIL-CIN1 | 56 (38/68) |
| LSIL-CIN2 | 81 (26/32) |
| LSIL-CIN3 | 80 (4/5) |
| HSIL-CIN2 | 78 (14/18) |
| HSIL-CIN3 | 86 (24/28) |
Discussion
The molecular mechanisms that determine malignant transformation of initially dysplastic cervical cells still remain unclear. Infection of cervical epithelial cells with high-risk HPV represents a necessary factor in development of cervical cancer. More than 15 types of high-risk HPV are now considered oncogenic, with HPV16 being the most dangerous.21 Viral load, persistence, and viral integration into the host genome were shown to be factors impacting the outcome of the disease. The benefit of HPV screening assays is their high negative predictive value for the presence of dysplasia.6 At the same time, it seems that the HPV screening by itself is not sufficient in predicting malignant transformation of dysplastic cervical lesions, because only a fraction of HPV-infected women will progress from atypical or mild dysplasia to high-grade lesions and subsequently to cancer.
To further improve on the informative value of HPV, we designed a novel fluorescence in situ hybridization assay, which permits simultaneous detection of HPV infection and chromosomal changes that occur in cervical epithelial cells. It is known that HPV infection is necessary but not sufficient to progress to high-grade dysplasia and cancer. Although the causes of cervical carcinogenesis are not completely understood, the use of fluorescent probes to detect aneusomy in genomic regions that are sensitive to destabilization may provide an essential tool for identifying cells at risk of progression. In an internal study, we assessed FISH probes to 35 unique loci using 30 biopsy specimens that consisted of normal, dysplastic, and cancer cases (I. Sokolova, A. O’Hare, W. King, S. Sitailo, M. Song, F. Policht, and A. Algeciras-Schimnick, manuscript in preparation). After initial experimentation, this collection of probes was narrowed down to the eight probes with the highest frequency of occurrence [3q26, 8q24, 20q13, Xp22, 1p31, CEP15, 3p14 (loss), and 3p21 (loss)]. Subsequent experimentation on a new set of 100 biopsy cases revealed that the 3q26 and the 8q24 regions had the highest frequency of copy number gains in samples with high-grade dysplasia and cancer. Specifically, 3q26 probe was positive in 100% of cancer specimens, 90% of CIN3 specimens, 78% of CIN2 specimens, 26% of CIN1 specimens, and 0% of normal specimens. The 8q24 probe was positive in 100, 95, 96, 26, and 5% of cancer, CIN3, CIN2, CIN1, and normal cases, respectively. In agreement with our findings, literature reports describe chromosomal abnormalities in pre-cancerous and cancerous cervical lesions. They include gains of the q arm of chromosome 3 and amplification of the q arm of chromosome 20 and the 8q24 region surrounding c-myc.17,19,20,22 Low-level amplifications of oncogenes located in 3q26 (PIK3CA), 5p15.33 (TERT), 8q24 (c-myc), 11q13.3 (CCND1), and 17q21.2 (ERBB2) have been shown to be associated with the development of cervical carcinoma. Accumulation of copy number imbalances using the comparative genomic hybridization approach was demonstrated on 70 snap-frozen cervical squamous intraepithelial lesions.23 DNA flow cytometry analysis of 85 punch biopsies of HPV16-positive cervical lesions in the study of Melsheimer et al24 revealed a statistically significant trend of aneuploidy associated with increasing severity of dysplasia. The 8q24 region has also been shown to be a frequent site of HPV integration and is often amplified in cervical cell lines and cervical lesions.25,26,27,28,29 MYC genes are key regulators of cell proliferation, and enhanced expression of MYC genes promotes unrestricted proliferation and contributes to the genesis of most human tumors.30 The 3q26 chromosome region was proposed as an important marker for disease progression in several studies.16,18,31 Therefore, the combination of HPV detection and the capacity to evaluate chromosomal aberrations may present a new possibility for the detection of progression of cervical dysplasia.
A few limitations of the FISH assay have been encountered that are related to sample heterogeneity. Extremely low viral loads in cells (one copy per cell) can be difficult to detect. The combination of elevated background and very limited quantities of HPV can cause a sample to be incorrectly identified as HPV negative. Samples that contain very few HPV-positive cells may also pose a challenge to the FISH assay. There may be cases where the HPV-positive cells are not deposited on the slide and, therefore, cannot be detected by the assay. The absence of HPV in very high-grade/cancer lesions may also occur because of uncontrolled cell proliferation and possible loss of HPV DNA from these cells. Although HPV would not be detected in these cells, the chromosome probes would detect the varying levels of aneusomy in the cells. In these cases, cells would probably exhibit very high levels of chromosomal abnormality, yet HPV would not be visible. It is important to note that these limitations occurred infrequently and need to be evaluated further to determine the extent of their impact on test sensitivity. The execution of further studies will help identify and eliminate deficiencies and provide more data to increase confidence in the assay, which may have a very significant clinical application.
From the clinical perspective, we hypothesize that this assay might be useful to stratify HPV-positive patients with an ASCUS or LSIL diagnosis before and/or after colposcopy. The presence of genomic instability in HPV-positive cells assessed by amplification of 3q26 or 8q24 chromosomal biomarkers may distinguish patients with clinically significant cervical lesions from those that are insignificant, ie, lesions with a high risk of progression from those with a low risk of progression. For example, an HPV-positive patient with a diploid 3q26/8q24 genotype should be classified as low risk of progression. Conversely, an HPV-positive patient with a 3q26/8q24 amplified genotype should be classified as high risk of progression. However, a larger controlled study with sufficient follow-up will be required to further evaluate the utility of this test and to compare it with currently used test methods (PCR and Hybrid Capture II) in the clinical management of women with HPV infection.
Note Added in Proof
After this article was accepted for publication, Kindt et al (32) reported on the role of survivin in toxin-induced acute renal failure.
Figure 3.
Average quantity of HPV-positive cells, chromosomally abnormal cells, and double-positive cells per histological category.
Acknowledgments
We are grateful to Dr. Carolyn Muller (University of New Mexico, Albuquerque, NM), Dr. Raheela Ashfaq (University of Texas, Southwest Medical Center, Dallas, TX), Dr. Nancy Kiviat (University of Washington, Seattle, WA), Dr. Elizabeth Unger (Centers for Disease Control and Prevention, Atlanta, GA), and Dr. Marek Skacel and Dr. Ray Tubbs (Cleveland Clinic Foundation, Cleveland, OH) for providing materials for the study and for valuable discussions.
Footnotes
Supported by a grant from Abbott Molecular (to K.C.H.).
Current address of A.A.-S.: Mayo Clinic, Department of Clinical Biochemistry and Immunology, Rochester, MN.
Current address of A.R.: Prescott Medical Communications, Chicago, IL.
Current address of W.K.: Whatman, Sanford, ME.
References
- Schiffman MH, Bauer HM, Hoover RN, Glass AG, Cadell DM, Rush BB, Scott DR, Sherman ME, Kurman RJ, Wacholder S, Stanton CK, Manos MM. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- Wright TC, Jr, Schiffman M. Adding a test for human papillomavirus DNA to cervical-cancer screening. N Engl J Med. 2003;348:489–490. doi: 10.1056/NEJMp020178. [DOI] [PubMed] [Google Scholar]
- Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32(Suppl 1):S16–S24. doi: 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Solomon D, Schiffman M, Tarone R. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J Natl Cancer Inst. 2001;93:293–299. doi: 10.1093/jnci/93.4.293. [DOI] [PubMed] [Google Scholar]
- Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. J Natl Cancer Inst. 2000;92:397–402. doi: 10.1093/jnci/92.5.397. [DOI] [PubMed] [Google Scholar]
- Wright TC, Jr, Schiffman M, Solomon D, Cox JT, Garcia F, Goldie S, Hatch K, Noller KL, Roach N, Runowicz C, Saslow D. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol. 2004;103:304–309. doi: 10.1097/01.AOG.0000109426.82624.f8. [DOI] [PubMed] [Google Scholar]
- Melnikow J, Nuovo J, Willan AR, Chan BK, Howell LP. Natural history of cervical squamous intraepithelial lesions: a meta-analysis. Obstet Gynecol. 1998;92:727–735. doi: 10.1016/s0029-7844(98)00245-2. [DOI] [PubMed] [Google Scholar]
- Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer. 2004;109:157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- Duensing S, Munger K. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J Virol. 2003;77:12331–12335. doi: 10.1128/JVI.77.22.12331-12335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing S, Munger K. Centrosome abnormalities and genomic instability induced by human papillomavirus oncoproteins. Prog Cell Cycle Res. 2003;5:383–391. [PubMed] [Google Scholar]
- Skyldberg B, Fujioka K, Hellstrom AC, Sylven L, Moberger B, Auer G. Human papillomavirus infection, centrosome aberration, and genetic stability in cervical lesions. Mod Pathol. 2001;14:279–284. doi: 10.1038/modpathol.3880303. [DOI] [PubMed] [Google Scholar]
- Münger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32(Suppl 1):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Kurtycz D, Nunez M, Arts T, Bauman C, Harris C, Inhorn S, Meisner L. Use of fluorescent in-situ hybridization to detect aneuploidy in cervical dysplasia. Diagn Cytopathol. 1996;15:46–51. doi: 10.1002/(SICI)1097-0339(199607)15:1<46::AID-DC9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Heselmeyer K, Macville M, Schrock E, Blegen H, Hellstrom AC, Shah K, Auer G, Ried T. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–240. [PubMed] [Google Scholar]
- Thein A, Trkova M, Fox M, Parrington J. The application of comparative genomic hybridization to previously karyotyped cervical cancer cell lines. Cancer Genet Cytogenet. 2000;116:59–65. doi: 10.1016/s0165-4608(99)00108-9. [DOI] [PubMed] [Google Scholar]
- Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in-situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer. 1999;24:144–150. doi: 10.1002/(sici)1098-2264(199902)24:2<144::aid-gcc7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Shera KA, McDougall JK. Genomic changes and HPV type in cervical carcinoma. Proc Soc Exp Biol Med. 2000;223:316–321. doi: 10.1046/j.1525-1373.2000.22345.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Schewe C, Petersen S, Salcedo M, Gariglio P, Schluns K, Dietel M, Petersen I. Human papilloma virus status and chromosomal imbalances in primary cervical carcinomas and tumour cell lines. Eur J Cancer. 2000;36:542–548. doi: 10.1016/s0959-8049(99)00323-8. [DOI] [PubMed] [Google Scholar]
- Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, Rush BB, Glass AG, Schiffman M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME, Ried T. Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol. 2003;163:1405–1416. doi: 10.1016/S0002-9440(10)63498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Maner S, Betz R, Angstrom T, Stendahl U, Bergman F, Zetterberg A, Wallin KL. Genetic alterations in cervical carcinomas: frequent low-level amplifications of oncogenes are associated with human papillomavirus infection. Int J Cancer. 2002;101:427–433. doi: 10.1002/ijc.10627. [DOI] [PubMed] [Google Scholar]
- Melsheimer P, Vinokurova S, Wentzensen N, Bastert G, von Knebel Doeberitz M. DNA aneuploidy and integration of human papillomavirus type 16 e6/e7 oncogenes in intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix uteri. Clin Cancer Res. 2004;10:3059–3063. doi: 10.1158/1078-0432.ccr-03-0565. [DOI] [PubMed] [Google Scholar]
- Macville M, Schrock E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, Popescu N, Ried T. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
- Mark HF. Fluorescence in-situ hybridization analysis of biomarkers in cancer. Exp Mol Pathol. 1999;67:131–134. doi: 10.1006/exmp.1999.2279. [DOI] [PubMed] [Google Scholar]
- Mark HF, Feldman D, Samy M, Sun C, Das S, Mark S, Lathrop J. Assessment of chromosome 8 copy number in cervical cancer by fluorescent in-situ hybridization. Exp Mol Pathol. 1999;66:157–162. doi: 10.1006/exmp.1999.2256. [DOI] [PubMed] [Google Scholar]
- Yu T, Ferber MJ, Cheung TH, Chung TK, Wong YF, Smith DI. The role of viral integration in the development of cervical cancer. Cancer Genet Cytogenet. 2005;158:27–34. doi: 10.1016/j.cancergencyto.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Peter M, Rosty C, Couturier J, Radvanyi F, Teshima H, Sastre-Garau X. MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene. 2006;25:5985–5993. doi: 10.1038/sj.onc.1209625. [DOI] [PubMed] [Google Scholar]
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt N, Menzenbach A, Van de Wouwer M, Betz I, De Vriese A, Conway EM. Protective role of the inhibitor of apoptosis protein, survivin, in toxin-induced renal failure. FASEB J. 2007 doi: 10.1096/fj.07-8882com. [Eup ahead of print] [DOI] [PubMed] [Google Scholar]