Abstract
Ser910 of FAK (focal adhesion kinase) was phosphorylated in fibroblasts treated with the phorbol ester PMA and dephosphorylated by PP1δ (protein phosphatase 1δ), as indicated by shRNA (small-hairpin RNA) gene silencing. Ser910 of FAK was reported previously to be an ERK (extracellular-signal-regulated kinase) 1/2 target in cells treated with phorbol esters. In contrast, various approaches, including the use of the MEK (mitogen-activated protein kinase/ERK kinase) inhibitors UO126 and CI-1040 to inhibit ERK1/2 pointed to the involvement of ERK5. This hypothesis was confirmed by: (i) shRNA ERK5 gene silencing, which resulted in complete pSer910 loss in non-stimulated and PMA-stimulated cells; (ii) direct phosphorylation of recombinant FAK by ERK5; and (iii) ERK5 activation by PMA. PMA stimulation and ERK5 silencing in MDA-MB 231 and MDA-MB 361 breast cancer cells indicated Ser910 targeting by ERK5 also in these cells. Given the proximity of Ser910 to the FAT (focal adhesion targeting) regulatory domain of FAK, cell proliferation and morphology were investigated in FAK−/− cells expressing S910A mutant FAK. The cell growth rate decreased and exposure to PMA induced peculiar morphological changes in cells expressing S910A, with respect to wild-type FAK, suggesting a role for Ser910 in these processes. The present study indicates, for the first time, the phosphorylation of Ser910 of FAK by ERK5 and its dephosphorylation by PP1δ, and suggested a role for Ser910 in the control of cell shape and proliferation.
Keywords: cell motility, cell survival, extracellular-signal-regulated kinase 5 (ERK5), focal adhesion kinase (FAK), protein phosphatase 1 (PP1), phorbol ester
Abbreviations: CAS, Crk-associated substrate; CDK, cyclin-dependent kinase; ERK, extracellular-signal-regulated kinase; FAK, focal adhesion kinase; FAT, focal adhesion targeting; FRNK, FAK-related non-kinase; GFP, green fluorescent protein; GSK3β, glycogen synthase kinase 3β; GST, glutathione transferase; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; PDB, phorbol 12,13-dibutyrate; PKC, protein kinase C; PP, protein phosphatase; shRNA, small-hairpin RNA; siRNA, small interfering RNA
INTRODUCTION
FAK (focal adhesion kinase) is a non-receptor tyrosine kinase that localizes to focal adhesions, the multimolecular structures of contact between cytoskeleton and extracellular matrix. FAK is activated upon cell attachment and spreading and is involved in diverse cellular processes, including cell migration, growth factor signalling and cell survival [1–4]. High FAK levels or deregulation of its signalling may contribute to tumour progression, invasiveness and metastasis [5–7].
FAK is phosphorylated on both tyrosine [1,2] and serine/threonine [1,8,9] residues. Some serine residues are located in close proximity to sites of protein–protein interaction, such as the paxillin-binding site in the FAT (focal adhesion targeting) domain and the proline-rich domain that binds the SH3 (Src homology 3) domain of CAS (Crk-associated substrate), suggesting their involvement in interactor binding and downstream signalling. Some of the serine residues display consensus sequences for proline-directed kinases and are potentially phosphorylated by members of the MAPK (mitogen-activated protein kinase) or CDK (cyclin-dependent kinase) families [8,9]. It was found that Ser722 is phosphorylated by GSK3β (glycogen synthase kinase 3β) during cell spreading and migration, and that phosphorylation modulates the FAK activity in a negative way [10]. Ser910 was reported to be phosphorylated by ERK (extracellular-signal-regulated kinase) 1/2 in cells treated with various agonists, including growth factors and phorbol ester PDB (phorbol 12,13-dibutyrate) [11,12]. Ser842 and Ser845 of FRNK (FAK-related non-kinase) (the C-terminal domain of FAK) were found phosphorylated in late mitosis, when FAK undergoes tyrosine-dephosphorylation and inactivation [13]. The proposed role for such phosphorylation is to dissociate FAK from other members of its signalling complex [14], when the focal adhesions are restructured [2] or disassembled in mitosis [15]. Dephosphorylation of these sites is less known. At mitotic exit, FAK serine-dephosphorylation occurs just before cell spreading in early G1 phase and involves PP (protein phosphatase) 1 [16]. The first indication of a link between FAK and PP1 [17] came from the immunofluorescence detection of PP1δ in focal adhesions [18,19]. Subsequently, it was found that PP1δ associated with FAK and FRNK in immunoprecipitation and pull-down assays [16]. PP1 is also responsible for Ser722 dephosphorylation in adherent cells [10] and at mitotic exit (E. Villa-Moruzzi, unpublished work).
ERK5, also known as big MAPK, is stimulated by a wide range of mitogens and cellular stresses, and contributes to regulate cell survival and proliferation, and cell differentiation [20–23]. The fact that ERK5 and ERK1/2 often act in synergy and that there is a lack of inhibitors that block ERK5 without affecting ERK1/2 suggest that ERK5 might regulate cellular functions formerly attributed to ERK1/2 [22]. ERK5 is constitutively activated in some tumour cells that overexpress ErbB and contributes to cell proliferation [24]. ERK5 was also reported to be involved, together with ERK1/2, in cytoskeletal disruption, suggesting its potential relevance to invasion mechanisms [25]. However, the specific ERK5 targets that mediate several of these processes are still unknown.
Owing to the proposed relevance of serine phosphorylation to FAK modulation, fibroblasts were treated with the phorbol ester PMA, a PKC (protein kinase C) activator known to affect both FAK and the cytoskeleton, and investigated the changes in the phosphorylation of FAK at Ser910 involving the kinase and phosphatase. Although the reported increase in pSer910 was confirmed, the present study revealed that Ser910 phosphorylation was due to ERK5 and not to ERK1/2, under both basal conditions and PMA stimulation. This is the first report of the involvement of ERK5 in the regulation of FAK.
MATERIALS AND METHODS
Materials
The antibodies to detect FAK (C-20), pTyr576/577 of FAK and GFP (green fluorescent protein) were from Santa Cruz Biotechnology. The antibodies to detect pSer722, pSer910 and pTyr397 of FAK were from BioSource. The anti-ERK5 antibodies were from Cell Signaling Technology. The anti-pERK5 antibodies (pThr218/pTyr220 of ERK5) were from Upstate. The anti-ERK1/2, anti-pERK1/2 and anti-α-tubulin antibodies were from Sigma–Aldrich. The anti-GST (glutathione transferase) antibodies and glutathione–Sepharose beads were from Amersham Biosciences. The MEK (MAPK/ERK kinase) 1/2 inhibitor UO126, the p38 inhibitor SB203580, the CDK inhibitor roscovitine and okadaic acid (K salt) were from Calbiochem. The MEK1/2 inhibitor CI-1040 was a gift from Pfizer. Tissue-culture and bacterial media, and additives, PMA and protease inhibitors were from Sigma–Aldrich. The PP1 catalytic subunit was purified from rabbit muscle as described in [26]. One unit of PP1 releases 1 nmol of H3PO4/min.
Cell culture, treatments and lysates
Fisher rat fibroblasts and NIH 3T3 cells were grown in DMEM (Dulbecco's modified Eagle's medium) added with 6% (v/v) fetal calf serum. MDA-MB breast cancer cells were grown in RPMI 1640 medium with 10% fetal calf serum added [16]. Subconfluent cells were exposed to PMA or okadaic acid or kinase inhibitors (all diluted in DMSO) or to DMSO alone (control), under the conditions specifically indicated. Following two washes in cold PBS, cells were lysed in 50 mM Tris/HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100 and 7.5 mM 2-mercaptoethanol (lysis buffer), plus 1 mM sodium orthovanadate (unless when metabolic labelling was performed, Figures 1A and 1B), 50 mM NaF or 1 μM okadaic acid and protease inhibitors [0.02% benzamidine, 0.02% PMSF, 0.02% Tos-Phe-CH2Cl (‘TPCK’) (tosylphenylalanylchloromethane), 10 μg/ml soya bean trypsin inhibitor and 4 μg/ml leupeptin].
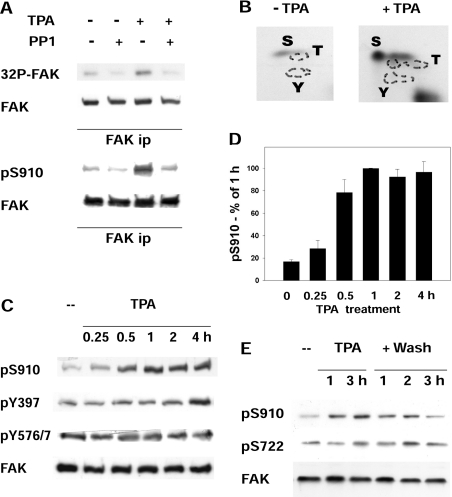
Figure 1. FAK phosphorylation and dephosphorylation in cells treated with PMA.
(A) Metabolic labelling of FAK and phosphorylation of Ser910. FAK was immunoprecipitated from fibroblasts that had been metabolically labelled with [32P]phosphoric acid and treated (+TPA) or not (−TPA) with 300 nM PMA for 4 h. The immunocomplexes were prepared in duplicate, and one complex from each treatment was incubated further with 2 units of muscle PP1 at 30 °C for 20 min (+PP1). Following SDS/8.5% PAGE and transblotting on to Immobilon-P, the membrane was first exposed to detect the radioactivity ([32P]FAK), then probed to detect FAK. In a similar experiment, but without metabolic labelling, FAK was immunoprecipitated (ip), treated with PP1 as above, and used to detect pSer910 of FAK (pS910) and FAK. (B) Phospho-amino acid analysis of labelled FAK. Two anti-FAK immunoprecipitates were prepared from [32P]phosphoric acid-labelled cells treated (+TPA) or not (−TPA) with PMA, as in (A). The FAK protein band was cut from the Immobilon-P membrane, hydrolysed at 110 °C for 1 h, analysed by two-dimensional high-voltage electrophoresis and exposed to detect the radioactivity. Residues are indicated using the single-letter amino acid code. (C) FAK phosphorylation at Ser910, Tyr397 and Tyr576/577. Cells were treated with 300 nM PMA (TPA) and collected at the indicated time points, and cell lysates were analysed by electrophoresis and immunoblotting, to detect pSer910 (pS910), FAK phosphorylation at Tyr397 (pY397) and Tyr576/577 (pY576/7), and FAK. (D) Quantification of the changes in Ser910 phosphorylation. The pSer910 bands from cells as in (C) were quantified by densitometric scanning and the data were normalized by adopting the 1 h value as 100%. Results are means±S.E.M. for four independent experiments. (E) FAK phosphorylation at Ser910 and Ser722 following PMA (TPA) treatment and subsequent PMA removal. PMA-treated cells as in (A) were either collected at 1 and 3 h, or washed twice at 3 h and incubated further for up to 3 h in the absence of PMA. Lysates from cells collected at the indicated time points were analysed to detect pSer722 of FAK (pS722), pSer910 (pS910) and FAK.
[32P]Pi metabolic labelling
The medium was changed into phosphoric acid-free medium added with 10% phosphoric acid-free fetal calf serum (previously dialysed against phosphoric acid-free medium). After 1 h, the medium was replaced with 1 ml of the same medium, added with [32P]phosphoric acid (Amersham Biosciences, 1 mCi/150 mm plate) and left for 5 h, with addition of 300 nM PMA after 1 h [16]. The extraction buffer contained 1 μM okadaic acid, to preserve serine and threonine phosphorylation, and no sodium orthovanadate, to favour FAK dephosphorylation on tyrosine (see Figure 1B and [16]).
Gene silencing
For transient PP1 or PP2A gene silencing, rat fibroblasts were transfected with isoform-specific rat siRNA (small interfering RNA) (Rn_Ppp1ca_1HP siRNA, Rn_Ppp1cc_1HP siRNA, Rn_Ppp1cb_3HP siRNA, Rn_Ppp2ca_1HP siRNA and Rn_Ppp2cb_1HP siRNA from Qiagen, targeting PP1α, PP1γ1, PP1δ, PP2Aα and PP2β respectively) or unrelated control siRNA, using HiPerFect Transfection reagent (Qiagen). The PP2A isoforms were targeted either separately or together (see Figure 2C). Cells were transfected at zero time, day 1 and day 2 (following 1:2 cell splitting). At day 3, cells were treated or not with 300 nM PMA for 1 h and lysed. For stable PP1δ gene silencing, NIH 3T3 cells were transfected with the pLKO.1-puro vector carrying the U6 promoter–mouse PP1δ (sense–linker–antisense) insert [constructs 97068915TRCN00000618 and 97068917TRCN00000620; Sigma Mission shRNA (small-hairpin RNA)] or the pLKO.1-puro empty vector, as control, using Lipofectamine™ 2000 transfection reagent (Invitrogen). Stable cell lines were established through selection with 5 μg/ml puromycin, starting from day 1 after transfection [10]. For ERK5 gene silencing, NIH 3T3 cells were co-transfected with the Banshee-GFP vector carrying the U6 promoter–mouse ERK5 (sense–linker–antisense) insert [23] or empty vector (both kindly supplied by Dr Astar Winoto and Dr Sue J. Sohn, Department of Molecular and Cell Biology, Division of Immunology and Cancer Research Laboratory, University of California, Berkeley, CA, U.S.A.) and the biCS2-puro vector. Transfection efficiency was tested by analysing the expression of the GFP. Cells were either lysed at day 3 after transfection or were grown further and subcloned by dilution in the presence of puromycin. MDA-MB 231 and MDA-MB 361 cells were transfected as above using the pLKO.1-puro vector carrying the U6 promoter–human ERK5 (sense–linker–antisense) insert (constructs TRCN0000010261 and TRCN000001354; Sigma Mission shRNA) or the pLKO.1-puro empty vector, and selected using puromycin, as above. Cells lysates were analysed by immunoblotting, and cells displaying total or subtotal gene silencing were grown further and used in the experiments. An α-tubulin immunoblot was used to quantify the extracts.
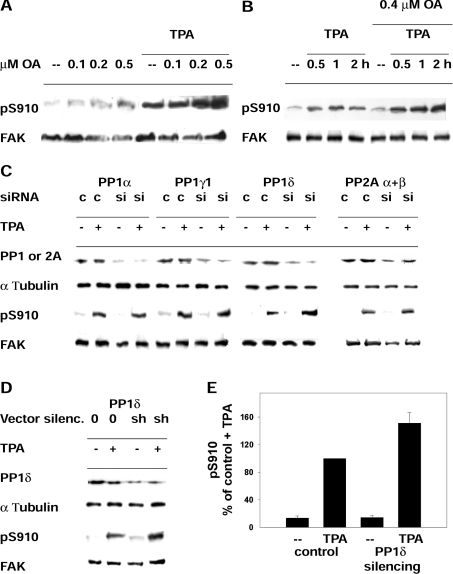
Figure 2. Phosphatase targeting of pSer910 of FAK.
(A) Phosphorylation of Ser910 in cells treated with PMA (TPA) and exposed further to increasing concentrations of okadaic acid (OA). Cells were incubated with or without 300 nM PMA for 40 min and with the indicated okadaic acid concentrations for an additional 40 min. Cells were collected, and lysates were analysed to detect pSer910 (pS910) and FAK. (B) Phosphorylation of Ser910 in cells treated with okadaic acid (OA) and exposed further to PMA (TPA). Cells were incubated with or without 0.4 μM okadaic acid for 20 min, subsequently with 300 nM PMA for up to 2 h and collected at the indicated time points. (C) Effect of transient silencing of PP1α, PP1γ1, PP1δ or PP2A (α+β) and of PMA (TPA) on pSer910. Rat fibroblasts were transfected with siRNA targeting the indicated PP1 or PP2A isoforms (si) or control siRNA (c) at zero time, day 1 and day 2, and collected at day 3, as described further in the Materials and methods section. PMA treatment (300 nM) before collecting cells was for 1 h. siRNA targeting PP2Aα and PP2Aβ were co-transfected. Following Western blotting, the membranes were probed to detect the PP1 or PP2A isoforms, α-tubulin, pSer910 (pS910) and FAK. Blots are representative of three independent experiments. (D) Effect of stable silencing (silenc.) of PP1δ and of PMA (TPA) on pSer910. NIH 3T3 fibroblasts were transfected with either shRNA vector targeting PP1δ (sh) or control empty vector (0), and stable transfectants were prepared by puromycin selection. Cell exposure to PMA and other reagents was as in (C). (E) Quantification of the effect of PMA on pSer910 in cells carrying stable PP1δ silencing. The pSer910 (pS910) bands from cells as in (D) were quantified by densitometric scanning, and the data obtained were normalized by adopting the control+PMA (TPA) value as 100%. Results are means±S.E.M. for four independent experiments, using two different siRNA constructs.
Wild-type and S910A mutant FAK stable transfectants
FAK−/− fibroblasts [4] were transfected with the pcDNA/HisMax TOPO vector expressing either wild-type or S910A mutant FAK, which also contains the Zeocin-resistance gene (see [10] for construct preparation and FAK−/− cells). Cells were exposed to 100 μg/ml Zeocin, starting from day 1 after transfection, grown for 2–3 weeks and subcloned by dilution in the presence of 40 μg/ml Zeocin. Clones expressing equal levels of wild-type and S910A FAK were used, and two clones for each cell-type were generally used to replicate the experiments.
Cell staining and proliferation assay
Cells were washed in PBS, fixed in 3% (w/v) parafomaldehyde and 2% (w/v) sucrose in PBS, pH 7.5, at room temperature (25 °C) for 10 min, washed in PBS and stained with Giemsa stain. For the proliferation assay, cells were fixed as above, stained with Crystal Violet and lysed in 10% (v/v) ethanoic (acetic) acid for measurement of absorbance at 595 nm (A595).
Bacterial growth, extracts and pull-down assays
Bacteria were grown, induced and extracted as described previously [16]. For pull-down assays, the GST-fusion proteins were bound to 25–50 μl of glutathione–Sepharose beads [16].
Immunoprecipitation and immunoblotting
Anti-FAK or anti-ERK5 antibodies were used for immunoprecipitation with Protein A–Sepharose [16]. Following incubation at 4 °C for 90 min with shaking and three washes in cell lysis buffer with 0.02% PMSF and 0.02% benzamidine, the immune complexes were boiled in Laemmli buffer. For PP1 treatment (see Figure 1A), the immune complexes were washed once more with 25 mM imidazole, pH 7.5, resuspended in 25 μl of the same buffer with 2 mM 2-mercaptoethanol, incubated with 2 units of muscle-purified PP1 catalytic subunit at 30 °C for 20 min and boiled in Laemmli buffer [16]. Electrophoresis was carried out on an SDS/8.5% polyacrylamide gel, and Immobilon-P membranes (Millipore) were used for transblotting. The membranes were probed with the indicated antibodies, followed by Protein A–horseradish peroxidase- or horseradish-peroxidase-conjugated secondary antibody (Sigma) and the ECL® (enhanced chemiluminescence) system (Amersham Biosciences). For re-probing with another antibody, the membranes were previously incubated in 5 mM phosphate buffer, 2% SDS and 2 mM 2-mercaptoethanol at 60 °C for 20 min.
Phospho-amino acid analysis
32P-labelled phosphoproteins, transferred to Immobilon-P, were hydrolysed in 6 M HCl at 110 °C for 1 h and analysed by two-dimensional high-voltage electrophoresis on thin-layer cellulose plates (Merck), followed by autoradiographic detection [27].
ERK5 activity assay
ERK5 was assayed using the ERK5-specific substrate Mef2. BL-21 Escherichia coli cells were transformed with the plasmid to express GST–Mef2C (kindly supplied by Dr Eric N. Olson, Department of Molecular Biology, Southwestern Medical Center, Dallas, TX, U.S.A.). The bacteria were cultured and extracted, and the Mef2–GST fusion protein was bound to glutathione–Sepharose beads as indicated above [16]. Following a final wash in 25 mM imidazole, pH 7.5, the beads were mixed with extract from cells that had been exposed or not to PMA and incubated for 5 min at 30 °C with shaking, in the presence of 10 mM MgCl2, 100 μM [γ-32P]ATP (2000–2500 c.p.m./pmol) (Amersham Biosciences) (modified from [10]). The reaction was terminated by adding Laemmli buffer and boiling. The 32P-labelled Mef2 was subjected to SDS/PAGE, followed by autoradiography. Subsequently, the Mef2 protein bands were excised from the gel and counted in a β-scintillation counter. Control assays, prepared with buffer instead of cell extract, were run in parallel, counted and subtracted from the c.p.m. of the assays.
Phosphorylation of Ser910 of FRNK
Wild-type or S910A mutant GST–FRNK [10] was bound to glutathione–Sepharose beads and incubated with immunoprecipitated ERK5 (obtained from fibroblasts exposed to 300 nM PMA for 1 h) in the presence of [γ-32P]ATP, 500 μM ATP and 10 mM MgCl2. The incubation was carried out at 30 °C for 30 min with shaking and was terminated by adding electrophoresis sample buffer.
RESULTS
Phosphorylation of FAK at Ser910 in cells treated with PMA
The phosphorylation and dephosphorylation of Ser910 (the homologue of chicken Ser911), which was reported to be phosphorylated in Swiss 3T3 cells treated with growth factors or the phorbol ester PDB [11,12], were analysed. FAK was immunoprecipitated from rat fibroblasts that had been metabolically labelled with [32P]phosphoric acid before and during exposure to the phorbol ester PMA (300 nM); this was followed by immunoblotting, autoradiography and phospho-amino acid analysis of FAK (Figures 1A and 1B). The results confirmed that PMA increased serine phosphorylation of FAK, specifically pSer910 (Figures 1A and 1B). pSer910 was analysed further in lysates of cells exposed to PMA for up to 4 h. The increase in pSer910 reached a maximum within 1 h (Figures 1C and 1D), whereas no major changes were detected at the FAK autophosphorylation Tyr397 site, or Src Tyr576/577 sites. Additionally, PMA did not affect the phosphorylation of Ser722 (Figure 1E), a site shown previously to be a GSK3β target during adhesion and migration [10].
The phosphatase targeting Ser910
Ser910 phosphorylation was reversed in vitro by PP1 (Figure 1A) and in the cell following PMA removal and further incubation without PMA (Figure 1E). To investigate which phosphatase targets Ser910 in the cell, the phosphatase inhibitor okadaic acid, which inhibits either PP2A alone or both PP2A and PP1, depending on the concentration [10,28], was used. Cells were incubated with or without PMA for 40 min and subsequently with increasing concentrations of okadaic acid for 40 min. In both untreated and PMA-treated cells, pSer910 increased only at 0.2 and 0.5 μM okadaic acid (Figure 2A), suggesting the involvement of PP1 rather than PP2A, generally inhibited at lower concentrations [28]. As a further investigation, cells were incubated with or without 0.4 μM okadaic acid for 20 min, and subsequently with PMA for up to 2 h. The increase in pSer910 in the presence of okadaic acid (Figure 2B) confirmed that a phosphatase, most likely PP1, was targeting pSer910 throughout the exposure to PMA.
To confirm the involvement of PP1, PP1 and PP2A were silenced in rat fibroblasts by transfecting siRNA targeting PP1α, PP1γ1, PP1δ or PP2Aα and PP2Aβ. The procedure adopted was a modification of the one used by Okada et al. [29] to silence PP1, and involved three subsequent siRNA transfections, one every 24 h, as described further in the Materials and methods section and legend to Figure 2. Silencing was achieved in the case of each gene, although often subtotal (see the Discussion and [29]). At 1 h before cell extraction, part of the cells was treated further with PMA. As expected, pSer910 levels were very low in untreated cells, and increased following PMA treatment (Figure 2C). However, silencing brought about a marked increase in pSer910 with respect to PMA-treated controls only in the case of PP1δ silencing (Figure 2C). This suggested the involvement of PP1δ in targeting pSer910, although the additional involvement of other phosphatases cannot be excluded completely (e.g. PP2A, owing to its incomplete silencing and a minor increase in pSer910 following PMA treatment).
To confirm the role of PP1δ, cells were prepared that carried stable gene silencing by transfecting a vector encoding for a PP1δ shRNA and selecting further by exposure to puromycin. Owing to the lack of a vector to silence the rat gene at the time of the experiments, mouse NIH 3T3 cells were used. Preliminary experiments indicated that PMA-dependent phosphorylation of Ser910 also occurred in these cells (results not shown). Two constructs, among those tested, targeting different PP1δ sequences, yielded similar silencing (see the Materials and methods section and the legend to Figure 2). Likewise, in the case of siRNA silencing, exposure to PMA increased pSer910 more in the cells carrying the stable silencing of PP1δ than in controls (Figures 2D and 2E). Altogether, the results indicated the involvement of PP1δ in targeting pSer910 and excluded the other phosphatases tested.
ERK1/2 is not involved in the PMA-stimulated phosphorylation of Ser910
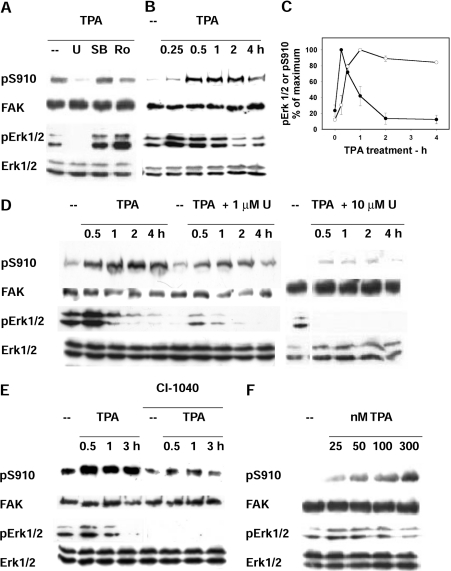
It has been reported that the activation of PKC by the phorbol ester PDB leads to phosphorylation of FAK at Ser910 by activated ERK1/2 [12]. Also in the system used in the present study, the use of the PKC inhibitors Go6983 and bisindolylmaleimide confirmed the involvement of PKC in the PMA-stimulated phosphorylation of FAK at Ser910 (results not shown). On the other hand, the present study led to results that argued against the involvement of ERK1/2, because of the following findings. First, the MEK1/2 inhibitor UO126 [30] indeed decreased pSer910, whereas neither the p38 inhibitor SB203580 nor the CDK inhibitor roscovitin did (Figure 3A); however, the time courses of ERK1/2 activation and of Ser910 phosphorylation were quite different. ERK1/2 activation was maximal at 15 min and subsequently declined (as judged by the changes in pERK1/2), whereas FAK phosphorylation at Ser910 peaked at 1 h and was sustained for up to 4 h (Figures 3B and 3C). Secondly, cell treatment with 1 μM UO126 (a concentration that inhibited ERK1/2 almost maximally; Figure 3D) resulted in a pSer910 loss that was much less pronounced than in the case of 10 μM UO126. Thirdly, incomplete inhibition of Ser910 phosphorylation was obtained also with another MEK1/2 inhibitor, CI-1040, under conditions that inhibited ERK1/2 completely (Figure 3E). Fourthly, the PMA concentrations required to activate ERK1/2 and to phosphorylate Ser910 were quite different. Ser910 was maximally phosphorylated at 300 nM PMA, whereas ERK1/2 was maximally activated at 25 nM PMA, and declined at higher concentrations (Figure 3F). In summary, the results suggested the involvement of a kinase that was different from ERK1/2.
Figure 3. Phosphorylation of FAK Ser910 and ERK1/2 activity in cells treated with PMA (TPA) and with kinase inhibitors.
(A) Treatment with kinase inhibitors. Cells were treated with 300 nM PMA for 1 h in the presence of the following kinase inhibitors: 10 μM U0126, to inhibit MEK1/2 (U); 10 μM SB203580, to inhibit p38 (SB) and 100 μM roscovitine, to inhibit CDKs (Ro). In addition to pSer910 (pS910) and FAK, the activation level of ERK1/2 (pERK1/2) and ERK1/2 protein were detected on cell lysates. (B) Time course of PMA treatment. Cells were treated with PMA for up to 4 h and analysed as in (A). (C) Quantification of the time course of PMA treatment. The pSer910 and pERK1/2 bands from blots as in (B) were quantified by densitometric scanning, and the data were normalized by adopting the maximal value obtained for each phosphoprotein as 100%. Results are means±S.E.M. for four independent experiments. (D) Treatment with U0126. Cells were treated with 300 nM PMA (TPA) in the presence or not of 1 or 10 μM U0126 (U). (E) Treatment with CI-1040. Cells were treated with 300 nM PMA in the presence or not of 1 μM of the MEK1/2 inhibitor CI-1040. (F) Treatment with increasing PMA concentrations. Cells were treated with the indicated PMA (TPA) concentrations for 1 h.
Gene silencing indicates the involvement of ERK5 in Ser910 phosphorylation, in both control and PMA-stimulated cells
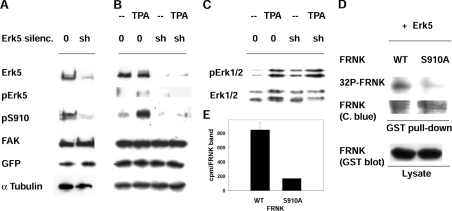
The MEK inhibitors used to inhibit ERK1/2 are known to also inhibit also ERK5, although with a higher IC50 [25,31]. To test the hypothesis of the involvement of ERK5, the gene was silenced and the consequences on pSer910 were investigated. Owing to the lack of a vector to silence the rat gene at the time of the experiments, mouse NIH 3T3 cells were used. Cells were transiently transfected with the ERK5 shRNA construct [23], or with the empty control vector, and the transfection efficiency was tested by analysing the level of the GFP, encoded by the same vector. To increase the system efficiency, cells were co-transfected with a puromycin-resistance vector, and the non-transfected cells (over 80%) were eliminated by exposure to puromycin [10]. Figure 4(A) shows the almost complete loss of ERK5 protein within 3 days, with respect to control cells. α-Tubulin levels confirmed the use of comparable amounts of cell extract. ERK5 silencing was accompanied by loss of pSer910 with respect to control cells (Figure 4A), thus indicating that ERK5 is the kinase that phosphorylates Ser910, or it is upstream to this site.
Figure 4. Involvement of ERK5 in the phosphorylation of FAK Ser910.
(A) Transient silencing (silenc.) of ERK5. NIH 3T3 cells were co-transfected with either empty vector (0) or the ERK5 shRNA vector (sh) together with a vector that expresses the puromycin resistance gene. At 24 h after transfection, the cells were exposed to 10 μg/ml puromycin, to kill non-transfected cells, and collected at 72 h. Cell lysates were used to detect ERK5, pSer910 (pS910), FAK, GFP (encoded by the transfected vectors) and α-tubulin. (B, C) PMA (TPA) treatment of cells carrying stable ERK5 silencing. Stable transfectants, obtained from cells transfected as in (A), were treated or not with 300 nM PMA for 1 h and analysed as in (A) (B) or for ERK1/2 activation (pERK1/2) and ERK1/2 protein (C). (D) Phosphorylation of Ser910 of FRNK by ERK5. GST–FRNK, either wild-type or S910A mutant, was bound to glutathione–Sepharose beads and incubated with immunoprecipitated ERK5 (obtained from fibroblasts exposed to 300 nM PMA for 1 h) in the presence of [γ-32P]ATP/MgCl2. This was followed by electrophoresis, Coomassie Blue staining and exposure to detect the radioactivity incorporated into FRNK (32P-FRNK). Wild-type and S910A-FRNK were detected also as GST-fusion proteins in the bacterial lysate used for the pull-down. (E) Quantification of [32P]FRNK (32P-FRNK). FRNK bands were excised from the gel (see D) and counted (c.p.m./FRNK band). Results are means±S.E.M. for three independent experiments. WT, wild-type.
To investigate the role of ERK5 in PMA-stimulated cells, stable transfectants, carrying either the silenced ERK5 gene or the control vector, were prepared from the puromycin-resistant clones. PMA stimulation did not lead to any change in Ser910 phosphorylation in cells that did not express ERK5, whereas pSer910 increased, as expected, in control cells (Figure 4B). The results clearly indicated that ERK5 also increased pSer910 following PMA stimulation. Additionally, ERK5 silencing did not affect ERK1/2 levels or the ability of PMA to stimulate ERK1/2 (Figure 4C), thus confirming that the silencing achieved was specific for ERK5.
Direct phosphorylation of Ser910 by ERK5
To test the ability of ERK5 to phosphorylate Ser910 directly, GST–FRNK, the GST-fusion protein of the C-terminal domain of FAK, was used. GST–FRNK, either wild-type or S910A mutant [10], was bound to glutathione–Sepharose beads and was mixed with ERK5 immunoprecipitated from PMA-treated fibroblasts. The reaction was carried out in the presence of [γ-32P]ATP and the 32P-labelled GST–FRNK was subjected to SDS/PAGE, autoradiography and counting in a β-scintillation counter. Wild-type FRNK was phosphorylated to a much higher level than was the S910A mutant (Figures 4D and 4E), indicating direct phosphorylation of Ser910 by ERK5.
ERK5 undergoes activation in cells stimulated with PMA
The next step was to assess the ability of PMA to activate ERK5. For this purpose, rat fibroblasts were exposed to PMA for up to 3 h, and the kinase activity of ERK5 was assayed with the specific substrate Mef2, used as GST–Mef2 bound to glutathione–Sepharose beads. The 32P-labelled Mef2 was subjected to SDS/PAGE, autoradiography and counting in a β-scintillation counter (Figures 5A and 5B). The results indicated that PMA activated ERK5, with a peak at 30 min and 1 h. Alternatively, ERK5 was immunoprecipitated from PMA-treated fibroblasts, and its phosphorylation at Thr218 and Tyr220, the hallmark of ERK5 activation, was probed (Figure 5C). Also this approach confirmed maximal ERK5 activation at 30 min and 1 h. Altogether, the results indicated ERK5 activation by PMA and the timescale supported its role in the phosphorylation of Ser910. However, ERK5 activation decreased subsequently, whereas pSer910 stayed high for up to 4 h (Figures 1 and 2). This discrepancy might be due to site targeting by a different kinase upon prolonged PMA stimulation or protection from dephosphorylation.
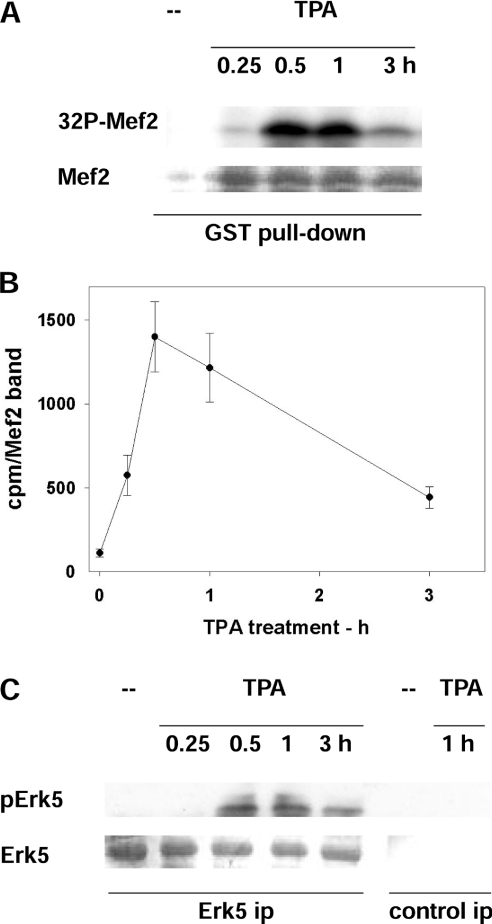
Figure 5. Activation of ERK5 in cells treated with PMA (TPA).
(A, B) ERK5 activity assayed as Mef2 kinase. (A) GST–Mef2 was bound to glutathione–Sepharose beads and incubated with [γ-32P]ATP/MgCl2 and cell extract from rat fibroblasts exposed to 300 nM PMA for the indicated time. This was followed by electrophoresis, Coomassie Blue staining (Mef2) and exposure to detect the radioactivity incorporated into Mef2 (32P-Mef2). (B) Quantification of 32P-Mef2. Mef2 bands were excised from the gel (see A) and counted (c.p.m./Mef2 band). Results are means±S.E.M. for three independent experiments. (C) ERK5 activity assayed as pERK5. ERK5 was immunoprecipitated (ERK5 ip), followed by electrophoresis, probing with antibodies that recognize the phosphorylation of Thr218 and Tyr220 of ERK5 (pERK5) and membrane staining with Coomassie Blue (ERK5). Control immunoprecipitates (control ip) are also shown.
Differential phosphorylation of Ser910 in MDA-MB 231 and MDA-MB 361 cells
To investigate the correlation between ERK5 and pSer910 in human cells, MDA-MB 231 and MDA-MB 361 breast cancer cells were adopted. pSer910 levels were high in non-stimulated MDA-MB 231 cells and were influenced little by PMA, whereas in MDA-MB 361 cells, pSer910 levels were very low and increased upon PMA stimulation (Figure 6A). Likewise, in the case of fibroblasts, the changes in pSer910 did not parallel ERK1/2 activation (Figure 6A).
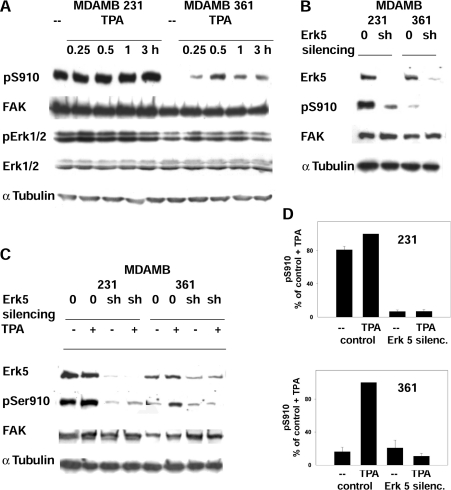
Figure 6. Role of PMA and ERK5 in the phosphorylation of FAK Ser910 in MDA-MB 231 and MDA-MB 361 cells.
(A) Phosphorylation of Ser910 and ERK1/2 activity in cells treated with PMA (TPA). Cells were treated with 300 nM PMA and collected at the indicated time points. Lysate analysis was as in Figure 4. (B) Phosphorylation of Ser910 following ERK5 silencing. Stable ERK5 silencing was obtained from cells transfected with either the empty vector (0) or the ERK5 shRNA vector (sh). (C) Phosphorylation of Ser910 following PMA (TPA) treatment of cells carrying silenced ERK5. Cells were treated with 300 nM PMA for 1 h and analysed as in (B). (D) Quantification of the effect of PMA on pSer910 (pS910) in cells carrying ERK5 silencing (silenc.). The pSer910 bands from cells in (C) were quantified by densitometric scanning and the data obtained were normalized by adopting the control+PMA value as 100%. Results are means±S.E.M. for four independent experiments, using two different siRNA constructs.
Gene silencing was used to test the involvement of ERK5. Cells were transfected with the shRNA construct targeting human ERK5 or with the empty vector, and stable transfectants were generated following exposure to puromycin. ERK5 silencing by two constructs targeting different ERK5 sequences (see the Materials and methods section) was accompanied by the loss of pSer910 in both cell lines (Figures 6B and 6D), suggesting ERK5 as the kinase that phosphorylates Ser910. As a further test, both stable transfectants and control cells were treated with PMA. Stimulation of ERK5-silenced cells led to little or no change in pSer910, whereas the pSer910 of control cells increased in MDA-MB 361 and to some extent also in MDA-MB 231 cells (Figures 6C and 6D). The results supported further the role of ERK5 in the PMA-stimulated phosphorylation of Ser910 in both cell lines.
Differential activation of ERK5 in MDA-MB 231 and MDA-MB 361 cells
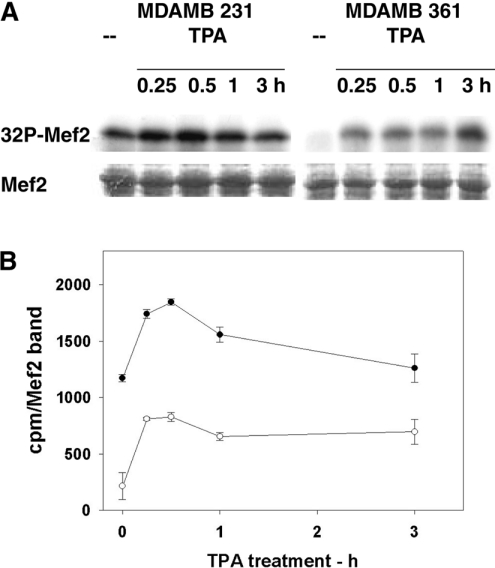
As in the case of fibroblasts (Figure 5), ERK5 activation following PMA treatment was investigated in MDA-MB 231 and MDA-MB 361 cells by assaying the Mef2 kinase activity. The PMA-induced ERK5 activation and increase in pSer910 were remarkably parallel in each cell line (Figures 7A and 7B). In MDA-MB 231 cells, both pSer910 and ERK5 activity displayed high basal values and little PMA stimulation (Figures 6D and 7B), whereas both pSer910 and ERK5 displayed low values and high PMA stimulation in MDA-MB 361 cells. These parallel changes supported further the hypothesis that ERK5 also phosphorylates Ser910 in these cells.
Figure 7. Activation of ERK5 in MDA-MB 231 and MDA-MB 361 cells treated with PMA (TPA).
ERK5 activity was assayed as Mef2 kinase. (A) Cells were treated and analysed as in Figure 5(A). (B) Quantification of 32P-labelled Mef2 (32P-Mef2). Mef2 bands were excised from the gel (see A) and counted (c.p.m./Mef2 band). Results are means±S.E.M. for three independent experiments.
Proliferation- and PMA-induced morphological changes in FAK−/− cells expressing wild-type or S910A mutant FAK
To investigate the biological functions of Ser910, FAK−/− fibroblasts in which FAK, either wild-type or S910A mutant, had been reintroduced were adopted. Stable transfectants were prepared using constructs described previously [10], and the mutation was confirmed by the lack of pSer910 following PMA stimulation (Figure 8A). As described previously, the mutation did not affect FAK activity or cell migration in a scratch-wound assay [10], and pSer910 did not change during cell spreading on fibronectin [10]. All of this suggested that the potential role of Ser910 was most likely to be unrelated to FAK regulation during the remodelling of focal adhesions that occurs in migration. On the other hand, considering that ERK5 is controlled by proliferation stimuli and that the role of FAK in cell survival involves the FAT domain of FAK, encompassing Ser910, it was tested whether the S910A mutation affected cell proliferation. For this purpose FAK−/− cells or FAK−/− cells expressing wild-type or S910A mutant FAK were plated at four different dilutions and tested for cell growth. The summary results (Figure 8B) indicated that cell growth was delayed in the cells expressing S910A mutant FAK, with respect to cells expressing wild-type FAK, suggesting a contribution from Ser910 to the role played by FAK in cell proliferation. Also, the mere reintroduction of FAK delayed cell growth (compare FAK−/− cells and cells expressing wild-type FAK in Figure 8B).
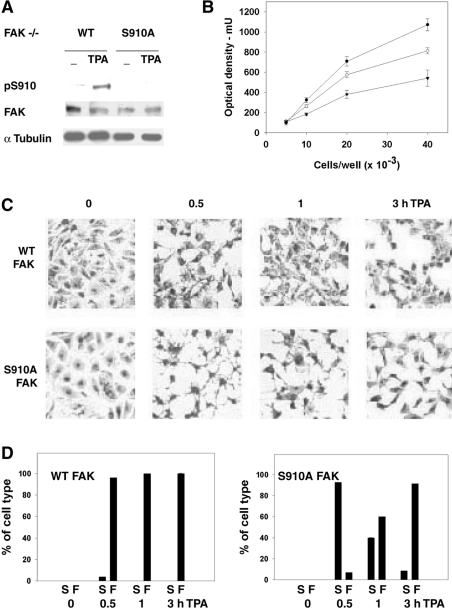
Figure 8. Effects of the expression of S910A FAK and PMA treatment in FAK−/− cells.
(A) Phosphorylation of Ser910 in cells treated with PMA (TPA). Cells were treated with 300 nM PMA for 1 h and cell lysates were analysed for pSer910 (pS910), FAK and α-tubulin. (B) Proliferation of cells expressing wild-type or S910A mutant FAK. The indicated amounts of FAK−/− cells expressing wild-type (○) or S910A FAK (▼) or control FAK−/− cells (●), were seeded in 24-well Costar plates, grown for 48 h, fixed, stained with Crystal Violet and analysed spectrophotometrically. Results are means±S.E.M. for five experiments, using two different clones for each cell type (three experiments in the case of FAK−/− cells). (C) Morphological changes in cells treated with PMA (TPA). Cells treated with PMA as in (A) were fixed at the indicated time points and stained with Giemsa stain. The images shown are representative of several experiments performed. (D) Quantification of the morphological changes in cells treated with PMA (TPA). Equivalent fields, displaying cells treated as in (C) (approx. 20–25 cells/field), were analysed for the presence of cells with retracted cytoplasm and long thin extensions (defined as star-like cells, S) and cells with fibroblast shape (defined as fibroblast-like, F). Results are percentages of S and F cells counted in each field and are means±S.E.M. for ten fields from three independent experiments. WT, wild-type.
PMA induces morphological changes that reflect cytoskeletal rearrangements [32]. In the present case, within 30 min, cells expressing wild-type FAK looked to be retracted, and cell protrusions appeared, leading to transition from a more polygonal (zero time) to a fibroblast-like shape (Figures 8C and 8D). In cells expressing S910A mutant FAK, retraction was more pronounced: cell protrusions were longer, thinner and more abundant, leading to a star-like morphology which prevailed at 30 min, was still detected at 1 h and had almost disappeared by 3 h (Figures 8C and 8D). In summary, the PMA-induced morphological changes were markedly different whether or not Ser910 was present, suggesting a role for Ser910 in regulating cell shape.
DISCUSSION
The phosphorylation of FAK at multiple tyrosine and serine sites modulates its activity and the association with signalling proteins. However, the kinases and phosphatases that target several of the serine residues are still unknown. The present study describes the targeting of Ser910 by ERK5 and PP1δ, and suggests a role for Ser910 in the regulation of cell proliferation and cell shape.
ERK5 responds to growth factors and stress stimuli, and contributes to differentiation and survival of normal and tumour cells [20–23]. ERK5 has been linked also to cytoskeletal disruption, with potential relevance to invasion mechanisms [25]. However, one problem that has hampered ERK5 studies is the difficulty of differentiating between ERK5 and ERK1/2 functions, since the two enzymes may act in synergy and share substrates, and also the inhibitors used in cellular studies are the same. Moreover, the roles played by ERK5 may vary depending on the cell type and the specific ERK5 targets are mostly unknown. In the present study, it was found that the phosphorylation of FAK at Ser910, previously attributed to ERK1/2, was due instead to ERK5, at least in the cell types examined.
Experiments of the present study exclude the involvement of ERK1/2 in the phosphorylation of Ser910 of FAK, based on: (i) the different time-courses of ERK1/2 activation and Ser910 phosphorylation; (ii) the different PMA concentrations required to activate ERK1/2 and to phosphorylate Ser910; and (iii) the incomplete inhibition of Ser910 phosphorylation induced by the MEK inhibitors UO126 and CI-1040, in the presence of total ERK1/2 inhibition. Subsequently, the positive involvement of ERK5 was indicated by: (i) ERK5 gene silencing, which resulted in complete loss of Ser910 phosphorylation in both unstimulated and PMA-stimulated fibroblasts, and (ii) activation of ERK5 in PMA-treated cells, assayed as both Mef2 kinase activity and pSer/pThr-ERK5 phosphorylation. Finally, the direct targeting of Ser910 by ERK5 was proven by the in vitro phosphorylation of Ser910 of FRNK. The conclusion differs from the one drawn by Hunger-Glaser et al. [11,12], indicating ERK1/2 as the kinase that phosphorylates Ser910 in Swiss 3T3 cells stimulated by various agonists, including PDGF (platelet-derived growth factor), FGF (fibroblast growth factor), EGF (epidermal growth factor), bombesin and the phorbol ester PDB. However, they also noticed incomplete inhibition of Ser910 phosphorylation in cells stimulated in the presence of low U0126 concentrations. On the other hand, both ERK1/2 [12] and ERK5 (the present study) did phosphorylate Ser910 of FRNK in vitro. Altogether, it cannot be excluded that, depending on the agonist and cell type, both ERK1/2 and ERK5 may target Ser910.
In MDA-MB 231 and MDA-MB 361 breast cancer cells, pSer910 displayed striking differences: it was very high in MDA-MB 231 cells, whereas it was almost undetectable in MDA-MB 361 cells. In both cell lines, ERK5 activity paralleled Ser910 phosphorylation, suggesting ERK5 involvement, and ERK5 gene silencing induced loss of pSer910, thus confirming Ser910 phosphorylation by ERK5. Considering the role of FAK in the survival of breast cancer cells [6] and the ability of Ser910 to favour cell proliferation (Figure 8B), Ser910 phosphorylation by ERK5 might contribute to cancer cell proliferation. In some breast cancer cell lines, ERK5 was reported to be part of the downstream signalling pathway of ErbB2 [24]. However, in MDA-MB 231 cells, high basal ERK5 activity was detected, in spite of low ErbB2. The lack of a positive correlation between ErbB2 and ERK5 might indicate that other activated receptors or oncogenes sustain ERK5 activity in these cells.
Several lines of evidence indicated PP1 as a major FAK serine phosphatase. Initially PP1δ was detected at focal adhesions and was found to associate with FAK [18,19]. Subsequently, it was found that PP1δ dephosphorylated FAK at mitotic exit [16] and was specifically targeting Ser722 in adherent cells [10]. Molecular analysis also indicated that the PP1δ domain interacting with FAK encompassed residues involved in catalysis [33]. The present data extended further the role of PP1δ in the dephosphorylation of Ser910. The procedure adopted to silence PP1 and PP2A included repetitive siRNA transfection and was described by Okada et al. [29] to silence PP1. In both investigations, silencing was not total, a situation that was probably favourable, since it allowed the cells to survive, in spite of interfering with proteins that regulate cell cycle [29]. Altogether, the data suggested a PP1δ contribution to focal adhesion stability by controlling FAK serine phosphorylation.
The role of FAK serine phosphorylation is still little understood. Previous work from my laboratory described FAK phosphorylation at Ser722 by GSK3β in cell spreading on fibronectin and during fibroblast migration [10]. Ser843 was phosphorylated by Ca2+/calmodulin-dependent protein kinase following stimulation with G-protein-coupled receptor agonists [34]. It has been suggested that serine phosphorylations contribute to disassemble the FAK signalling complex and prevent the interaction with CAS or paxillin during focal adhesion rearrangements in cell migration [2,5,8,35] and mitosis [15]. Conversely, FAK serine dephosphorylation was reported to stabilize adhesion in HT-29 colon carcinoma cells [36]. Altogether, these observations point to the importance of keeping the phosphorylation of specific serine residues under strict control, in order to preserve the stability of the FAK signalling complex.
In most cells, the proliferation response to growth factors depends on cell adhesion to the extracellular matrix. FAK plays a primary role in this process by stimulating various downstream signalling pathways, which depend on both FAK activity and complex formation with adaptor proteins, such as CAS and paxillin. By this means, growth factors and integrin signalling cooperate in controlling cell growth and progression through the cell cycle [1,2,5,6]. Several molecular details of this mechanism have still to be unravelled. The results of the present study suggest a role for Ser910, since its mutation delayed cell proliferation. Given the fact that ERK5 is regulated by growth factors and stress stimuli, and contributes to cell survival [20–23], it is conceivable that one of the survival pathways involves stimulation of ERK5 and FAK phosphorylation at Ser910.
The morphological changes induced by PMA were more profound and persistent in cells expressing S910A mutant FAK, suggesting a role for Ser910 in FAK downstream signalling to the cytoskeleton that regulates cell morphology. The involvement of ERK5 strongly supports this view, since also ERK5 has been linked to cytoskeletal disruption, through a mechanism that is not completely clear. The phosphorylation of Ser910 by ERK5 may contribute to fill this gap.
Unravelling the molecular interactions occurring downstream of Ser910 may be more problematic. The location of Ser910 within the FAT domain [1,2,5] suggested its involvement in the interaction with paxillin, a scaffold protein linking FAK to the actin cytoskeleton [14,37]. Additionally, paxillin is phosphorylated in a PKC-dependent way [14] and by ERK1/2 during focal adhesion disassembly [37], suggesting that the paxillin–FAK association might also be influenced by changes at Ser910. The finding that the S910A mutation increased paxillin association seemed to confirm this hypothesis [12]. On the other hand, preliminary data from my laboratory, not included in the present paper, did not support this view (E. Villa-Moruzzi, unpublished work). It was found that the increase in pSer910 induced by PMA was accompanied by an increase, rather than a decrease, in the FAK–paxillin association, whereas loss of Ser910 phosphorylation (owing to ERK5 silencing) did not significantly affect the PMA-stimulated association. On the basis of these discrepancies, it is suggested that the potential role of Ser910 in the regulation of the FAK–paxillin association may be more complex and its unravelling requires deeper investigation.
Acknowledgments
This work was supported by PRIN (Progetti di Ricerca di Interesse Nazionale) and FIRB (Fondo Investimenti Ricerca di Base) grants from MIUR (Ministero dell'Istruzione, dell'Università e della Ricerca Scientifica) (Rome) and the University of Pisa. I thank Dr Mariarita Bianchi (Department of Experimental Pathology, University of Pisa) for performing the experiment shown in Figure 3(A), Dr Astar Winoto and Dr Sue J. Sohn for supplying the plasmid to silence mouse ERK5, Dr Eric N. Olson for supplying the GST–Mef2 construct, Pfizer for supplying the MEK inhibitor CI-1040 and Dr Rony Seger (Weizmann Institute of Science, Rehovot, Israel) for discussions and fruitful suggestions.
References
- 1.Hanks S. K., Ryzhova L., Shin N., Brabek J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front. Biosci. 2003;8:d982–d986. doi: 10.2741/1114. [DOI] [PubMed] [Google Scholar]
- 2.Parsons J. T. Focal adhesion kinase: the first ten years. J. Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 3.Carragher N. O., Frame M. C. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 2004;14:241–249. doi: 10.1016/j.tcb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Owen J. D., Ruest P. J., Fry D. W., Hanks S. K. Induced focal adhesion kinase (FAK) expression in FAK-null cells enhances cell spreading and migration requiring both auto- and activation loop phosphorylation sites and inhibits adhesion-dependent tyrosine phosphorylation of Pyk2. Mol. Cell. Biol. 1999;19:4806–4818. doi: 10.1128/mcb.19.7.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlaepfer D. D., Mitra S. K., Ilic D. Control of motile and invasive phenotypes by focal adhesion kinase. Biochim. Biophys. Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Schatzmann F., Marlow R., Streuli C. H. Integrin signaling and mammary cell function. J. Mammary Gland Biol. Neoplasia. 2004;8:395–408. doi: 10.1023/B:JOMG.0000017427.14751.8c. [DOI] [PubMed] [Google Scholar]
- 7.Mitra S. K., Schlaepfer D. D. Integrin-regulated FAK–Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Ma A., Richardson A., Schaefer E. M., Parsons J. T. Serine phosphorylation of focal adhesion kinase in interphase and mitosis: a possible role in modulating binding to p130CAS. Mol. Biol. Cell. 2001;12:1–12. doi: 10.1091/mbc.12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigera P. R., Jeffery E. D., Martin K. K., Shabanowitz J., Hunt D. F., Parsons J. T. FAK phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 2005;118:4931–4935. doi: 10.1242/jcs.02696. [DOI] [PubMed] [Google Scholar]
- 10.Bianchi M., De Lucchini S., Marin O., Turner D. L., Hanks S. K., Villa-Moruzzi E. Regulation of FAK Ser722 phosphorylation and kinase activity by GSK3 and PP1 during cell spreading and migration. Biochem. J. 2005;391:359–370. doi: 10.1042/BJ20050282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunger-Glaser I., Fan R. S., Perez-Salazar E., Rozengurt E. PDGF and FGF induce focal adhesion kinase (FAK) phosphorylation at Ser-910: dissociation from Tyr-397 phosphorylation and requirement for ERK activation. J. Cell. Physiol. 2004;200:213–222. doi: 10.1002/jcp.20018. [DOI] [PubMed] [Google Scholar]
- 12.Hunger-Glaser I., Perez-Salazar E., Sinnett-Smith J., Rozengurt E. Bombesin, lysophosphatidic acid, and epidermal growth factor rapidly stimulate focal adhesion kinase phosphorylation at Ser-910. J. Biol. Chem. 2003;278:22631–22643. doi: 10.1074/jbc.M210876200. [DOI] [PubMed] [Google Scholar]
- 13.Richardson A., Shannon J. D., Adams R. B., Schaller M. D., Parsons J. T. Identification of integrin-stimulated sites of serine phosphorylation in FRNK, the separately expressed C-terminal domain of focal adhesion kinase: a potential role for protein kinase A. Biochem. J. 1997;324:141–149. doi: 10.1042/bj3240141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown M. C., Turner C. E. Paxillin: adapting to change. Physiol. Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 15.Yamakita Y., Totsukawa G., Yamashiro S., Fry D., Zhang X., Hanks S. K., Matsumura F. Dissociation of FAK/p130(CAS)/c-Src complexes during mitosis: role of mitosis-specific serine phosphorylation of FAK. J. Cell Biol. 1999;144:315–324. doi: 10.1083/jcb.144.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fresu M., Bianchi M., Parsons J. T., Villa-Moruzzi E. Cell-cycle-dependent association of protein phosphatase 1 and focal adhesion kinase. Biochem. J. 2001;350:407–414. doi: 10.1042/0264-6021:3580407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceulemans H., Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 18.Murata K., Hirano K., Villa-Moruzzi E., Hartshorne D. J., Brautigan D. L. Differential localization of myosin and myosin phosphatase subunits in smooth muscle cells and migrating fibroblasts. Mol. Biol. Cell. 1997;8:663–673. doi: 10.1091/mbc.8.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villa-Moruzzi E., Tognarini M., Cecchini G., Marchisio P. C. Protein phosphatase 1δ is associated with focal adhesions. Cell Adhes. Commun. 1998;5:297–305. doi: 10.3109/15419069809040299. [DOI] [PubMed] [Google Scholar]
- 20.Raman M., Cobb M. H. MAP kinase modules: many roads home. Curr. Biol. 2003;13:R886–R888. doi: 10.1016/j.cub.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 21.Nishimoto S., Nishida E. MAPK signaling: ERK5 versus ERK1/2. EMBO Rep. 2003;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Tournier C. Regulation of cellular functions by the ERK5 signalling pathway. Cell. Signalling. 2006;18:753–760. doi: 10.1016/j.cellsig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Sohn S. J., Li D., Lee L. K., Winoto A. Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol. Cell. Biol. 2005;25:8553–8566. doi: 10.1128/MCB.25.19.8553-8566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esparis-Ogando A., Diaz-Rodriguez E., Montero J. C., Yuste L., Crespo P., Pandiella A. Erk5 participates in neuregulin signal transduction and is constitutively active in breast cancer cells overexpressing ErbB2. Mol. Cell. Biol. 2002;22:270–285. doi: 10.1128/MCB.22.1.270-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barros J. C., Marshall C. J. Activation of either ERK1/2 or ERK5 MAP kinase pathways can lead to disruption of the actin cytoskeleton. J. Cell Sci. 2005;118:1663–1671. doi: 10.1242/jcs.02308. [DOI] [PubMed] [Google Scholar]
- 26.Resink T. J., Hemmings B. A., Tung H. Y. L., Cohen P. Characterization of a reconstituted Mg-ATP-dependent protein phosphatase. Eur. J. Biochem. 1983;133:455–461. doi: 10.1111/j.1432-1033.1983.tb07485.x. [DOI] [PubMed] [Google Scholar]
- 27.Kamps P. M. Determination of phosphoamino acid composition by acid hydrolysis of protein blotted to Immobilon. Methods Enzymol. 1991;201:21–27. doi: 10.1016/0076-6879(91)01005-m. [DOI] [PubMed] [Google Scholar]
- 28.Schoenthal A. H. Analyzing gene expression with the use of serine/threonine phosphatase inhibitors. Methods Mol. Biol. 1998;93:35–40. doi: 10.1385/0-89603-468-2:35. [DOI] [PubMed] [Google Scholar]
- 29.Okada T., Fujii T., Tanuma N., Mitsuhashi S., Urano T., Araki Y., Shima H., Kikuchi K. Analysis of isoform specific function of PP1 catalytic subunit in mammalian cells using siRNA. Int. J. Oncol. 2004;25:1383–1388. [PubMed] [Google Scholar]
- 30.Schaul Y. D., Seger R. Use of inhibitors in the study of MAPK signaling. Methods Mol. Biol. 2004;250:113–125. doi: 10.1385/1-59259-671-1:113. [DOI] [PubMed] [Google Scholar]
- 31.Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 32.Keenan C., Kelleher D. Protein kinase C and the cytoskeleton. Cell. Signalling. 1998;10:225–232. doi: 10.1016/s0898-6568(97)00121-6. [DOI] [PubMed] [Google Scholar]
- 33.Bianchi M., De Lucchini S., Vietri M., Villa-Moruzzi E. Reciprocally interacting domains of protein phosphatase 1 and focal adhesion kinase. Mol. Cell. Biochem. 2005;272:85–90. doi: 10.1007/s11010-005-7639-z. [DOI] [PubMed] [Google Scholar]
- 34.Fan R. S., Jacamo R. O., Jang S., Sinnett-Smith J., Rozengurt E. G protein-coupled receptor activation rapidly stimulates focal adhesion kinase phosphorylation at Ser-843. J. Biol. Chem. 2005;280:24212–24220. doi: 10.1074/jbc.M500716200. [DOI] [PubMed] [Google Scholar]
- 35.Young M. R., Kolesiak K., Meisinger J. Protein phosphatase-2A regulates endothelial cell motility and both the phosphorylation and the stability of focal adhesion complexes. Int. J. Cancer. 2002;100:276–282. doi: 10.1002/ijc.10491. [DOI] [PubMed] [Google Scholar]
- 36.Haier J., Nicolson G. L. Time-dependent dephosphorylation through serine/threonine phosphatases is required for stable adhesion of highly and poorly metastatic HT-29 colon carcinoma cell lines to collagen. Anticancer Res. 2000;20:2265–2271. [PubMed] [Google Scholar]
- 37.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. FAK-Src signaling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]