Abstract
Arginine methylation of histone H3 and H4 plays important roles in transcriptional regulation in eukaryotes such as yeasts, fruitflies, nematode worms, fish and mammals; however, less is known in plants. In the present paper, we report the identification and characterization of two Arabidopsis thaliana protein arginine N-methyltransferases, AtPRMT1a and AtPRMT1b, which exhibit high homology with human PRMT1. Both AtPRMT1a and AtPRMT1b methylated histone H4, H2A, and myelin basic protein in vitro. Site-directed mutagenesis of the third arginine (R3) on the N-terminus of histone H4 to lysine (H4R3N) completely abolished the methylation of histone H4. When fused to GFP (green fluorescent protein), both methyltransferases localized to the cytoplasm as well as to the nucleus. Consistent with their subcellular distribution, GST (glutathione transferase) pull-down assays revealed an interaction between the two methyltransferases, suggesting that both proteins may act together in a functional unit. In addition, we demonstrated that AtFib2 (Arabidopsis thaliana fibrillarin 2), an RNA methyltransferase, is a potential substrate for AtPRMT1a and AtPRMT1b, and, furthermore, uncovered a direct interaction between the protein methyltransferase and the RNA methyltransferase. Taken together, our findings implicate AtPRMT1a and AtPRMT1b as H4-R3 protein arginine N-methyltransferases in Arabidopsis and may be involved in diverse biological processes inside and outside the nucleus.
Keywords: Arabidopsis thaliana, arginine methylation, fibrillarin 2, histone modification, protein arginine N-methyltransferase (PRMT), RNA methyltransferase
Abbreviations: DART1, Drosophilaarginine methyltransferase 1; Fib, fibrillarin; AtFib2, Arabidopsis thaliana fibrillarin 2; FLC, FLOWERING LOCUS C; GAR, glycine- and arginine-rich; GFP, green fluorescent protein; GST, glutathione transferase; HMT1, heterogeneous nuclear ribonucleoprotein methyltransferase 1; MBP, maltose-binding protein; PRMT, protein arginine N-methyltransferase; AtPRMT, Arabidopsis thaliana PRMT; hPRMT, human PRMT; R3, third arginine residue; RBD, RNA-binding domain; SAM/AdoMet, S-adenosylmethionine; SKB1, Shk1 kinase-binding protein; snoRNP, small nucleolar ribonucleoprotein particle
INTRODUCTION
In the nuclei of eukaryotic cells, chromatin is the physiological template for various genetic processes. The basic unit of chromatin is the nucleosome, which is composed of an octamer of core histone proteins H2A, H2B, H3 and H4 (two copies of each), around which is wrapped 146 base pairs of DNA. Histones contain flexible N-terminal tails that undergo diverse post-translational modifications, including acetylation, phosphorylation, methylation, ubiquitination, ADP-ribosylation and SUMOylation [1]. The pattern of these modifications is proposed to constitute a ‘histone code’ [2]. During the last few years, the identification and characterization of multiple lysine and arginine methyltransferases has not only highlighted, but also shaped our understanding of post-translational modifications to a more extended level: the ‘protein code’ [3–8].
Although discovered several decades ago, studies on arginine methylation have flourished rather recently. From yeasts to mammals and plants, protein arginine methylation is involved in multiple cellular processes, including transcriptional regulation, DNA damage response, RNA processing, nuclear trafficking, signal transduction, formation of silent chromatin and cell fate determination [9–11]. The enzymes catalysing arginine methylation, PRMTs (protein arginine N-methyltransferases), transfer methyl groups from SAM/AdoMet (S-adenosylmethionine) to the guanidino nitrogens of arginine residues. All known active PRMTs in mammals catalyse the formation of L-NMMA (NG-monomethyl-L-arginine). These are classified as Type I or Type II enzymes on the basis of whether subsequent dimethylation is asymmetric [ADMA (asymmetric ω-NG,NG-dimethylarginine)] or symmetric [SDMA (symmetric ω-NG,N′G-dimethylarginine)] [12].
Currently, nine members of the PRMT family have been identified in mammals (designated PRMT1–PRMT9), among which PRMT1 and CARM1 (coactivator-associated arginine methyltransferase 1) represent the Type I PRMTs that possess asymmetric methyltransferase activities to histones H4 [8] and H3 [3] respectively, whereas PRMT5/SKB1 (Shk1 kinase-binding protein 1) represents the Type II PRMTs that deposit symmetric dimethyl groups on histones H2A, H3 and H4 [13,14]. The functional consequences of these histone modifications may correlate with either transcriptional activation [3,8,15] or repression [16].
PRMT1, the predominant methyltransferase that accounts for more than 85% Type I methyltransferase activity in mammalian cells and tissues [17], was initially identified as a TIS21 (PMA-inducible sequence 21)- and BTG1 (B-cell translocation gene 1)-interacting protein in yeast two-hybrid screens [18], and was subsequently shown as a nuclear receptor coactivator, which facilitated transcriptional activation by methylating histone H4 at the third arginine (R3) [8,15,19,20]. Since the growth of embryos from Prmt1−/− mice was arrested shortly after implantation, PRMT1 must play a critical role in early mouse development [21]. In addition to histone H4, a wide range of non-histone proteins have also been reported as substrates for PRMT1 or HMT1 (heterogeneous nuclear ribonucleoprotein methyltransferase 1; the major arginine methyltransferase in yeast), especially those RNA-binding proteins with RG (Arg-Gly)/RGG (Arg-Gly-Gly) or RXR (Arg-Xaa-Arg) motifs [19,22–29].
Although the roles of H4-R3 asymmetric dimethylation in transcriptional regulation have been well documented in mammals, their function remains largely unknown in plants. Recently, we and another group have independently reported the Arabidopsis PRMT5/SKB1-directed symmetric methylation on histone H4-R3 negatively regulated flowering time by repressing the transcription of FLC (FLOWERING LOCUS C) [30,31]. In the present paper, we report the identification and characterization of two Arabidopsis thaliana genes, AtPRMT1a and AtPRMT1b, both of which encode the plant homologues of hPRMT (human PRMT) 1. We demonstrate that AtPRMT1a and AtPRMT1b are bona fide Type I PRMTs, which asymmetrically methylate histone H4 at R3 and non-histone proteins such as the RNA methyltransferase AtFib2 (Arabidopsis thaliana fibrillarin 2). Our results suggest this pair of methyltransferases may play diverse roles in transcriptional regulation and RNA processing by methylating histones and the RNA methyltransferase AtFib2 respectively.
EXPERIMENTAL
Bioinformatic tools
Database searching was performed using TBLASTN at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple alignments of protein sequences were performed with ClustalX and were manually edited with the GeneDoc program.
DNA constructs
Full-length coding sequences of AtPRMT1a, AtPRMT1b, AtFib2, the N-terminal region of AtFib2 (AtFib2N1–150, corresponding to nucleotides 1–450 of AtFib2), the N-terminal region of histone H4 (H4R3N1–54, corresponding to nucleotides 1–162 of histone H4) and H4K3N1–54 (R3 mutated into lysine) were amplified by PCR. Primer pairs are listed as follows: CX0127 (5′-CGCGGATCCATGACTAGTACGGAGAACAACAACAAC-3′) and CX0128 (5′-CTCGAATTCTTAGCGCATCTTATAGAAGTGG-3′) for AtPRMT1a; CX0135 (5′-CGCGGATCCATGACTAAGAACAGTAACCACGACGAGAAT-3′) and CX0136 (5′-CTCGAATTCCTTCTGAAGTTCTGCTTCTTATG-3′) for AtPRMT1b; CX0364 (5′-CGCGGATCCATGAGACCTCCTCTAACTGGAAGTGGT-3′) and CX0365 (5′-CTCGAATTCACTTCTAAGCAGCAGTAGCAGC-3′) for AtFib2; CX0364 (5′-CGCGGATCCATGAGACCTCCTCTAACTGGAAGTGGT-3′) and CX0765 (5′-CTCGAATTCAGATGTTATCAACACCACCAAGA-3′) for AtFib2N1–150; CX0348 (5′-CGCGGATCCATGTCTGGGCGAGGTAAAGGTGGCAAG-3′) and CX0171 (5′-CTCGAATTCTCACTCCTCGTAGATGAGACCA-3′) for H4R3N1–54; and CX0170 (5′-CGCGGATCCATGTCTGGGAAAGGTAAAGGTGGCA-3′) and CX0171 (5′-CTCGAATTCTCACTCCTCGTAGATGAGACCA-3′) for H4K3N1–54.
These PCR fragments were digested further and cloned in-frame into the BamHI (GGATCC, underlined) and EcoRI (GAATTC, underlined) sites of pGEX-4T-2 vector (Amersham Biosciences) to generate the GST–AtPRMT1a, GST–AtPRMT1b, GST–AtFib2, GST–AtFib2N1–150, GST–H4R3N1–54 and GST–H4K3N1–54 GST (glutathione transferase) fusion constructs.
To generate the GFP–AtPRMT1a and GFP–AtPRMT1b GFP (green fluorescent protein) expression constructs, the GST–AtPRMT1a and GST–AtPRMT1b fusion constructs were digested with EcoRI and the cohesive ends were filled-in using Klenow fragment (NEB), then digested with BamHI. The resulting fragments of AtPRMT1a and AtPRMT1b were subcloned into the pAVA321 vector, which was digested using the same strategy, except for the last step, using BglII digestion instead of BamHI.
To generate an MBP (maltose-binding protein)-tagged AtFib2 fusion construct (MBP–AtFib2), fragments of AtFib2 were digested with BamHI and EcoRI from the GST–AtFib2 fusion construct and subcloned into pMal-2×His-TEV vector. To generate the MBP–AtPRMT1b fusion construct, fragments of AtPRMT1b were digested with SmaI and BamHI and reinserted into pMal-2×His-TEV vector.
All of the constructs generated above were verified by DNA sequencing.
Preparation of the fusion proteins and methylation assay
The fusion proteins GST–AtPRMT1a, GST–AtPRMT1b, GST–H4R3N1–54, GST–H4K3N1–54, GST–AtFib2, GST–AtFib2N1–150, and MBP–AtPRMT1a were expressed in Escherichia coli strain BL21(RIL) cells, and MBP–AtFib2 was expressed in E. coli strain BL21(DE3) cells, and all were affinity-purified with glutathione–Sepharose beads (Amersham Biosciences). In vitro histone methyltransferase assays were performed as described previously [32]. Labelled proteins were separated by SDS/15% PAGE and visualized by autofluorography for 48 h.
Isolation of histones from Arabidopsis
Approx. 3 g of leaves from 2-week-old Arabidopsis were homogenized in histone extraction buffer [0.25 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM Pipes, pH 6.8, 0.8% Triton X-100 and protease inhibitor cocktail (Roche)]. After centrifugation at 10000 g for 20 min, the pellet was resuspended with 0.4 M H2SO4, then centrifuged at 22000 g for 5 min. Then, 12 vol. of acetone were added to the supernatant and left at −20 °C overnight to precipitated proteins. The proteins were then centrifuged at 8000 g for 15 min and resuspended with 4 M urea to a final volume of 100 μl. All steps were carried out at 4 °C unless specified.
Western blot analysis
Recombinant histone H4, calf thymus histones, calf thymus histones methylated by AtPRMT1a and total histones isolated from Arabidopsis were separated by SDS/15% PAGE, transferred on to a PVDF membrane (Millipore) and probed with anti-(asymmetric dimethyl-H4R3) (anti-H4R3ame2) antibodies (Upstate) [1:1000 dilution in TBST (0.1% Tween 20, 150 mM NaCl and 50 mM Tris/HCl, pH 7.5)].
Subcellular localization
GFP–AtPRMT1a- and GFP–AtPRMT1b-expressing plasmids were transiently expressed in onion epidermal cells as described previously [33]. The inner epidermal layers from fresh onions (obtained from a local market) were placed on MS (Murashige and Shoog) basal medium. A 5 μg amount of plasmids containing the GFP–AtPRMT1a or GFP–AtPRMT1b fusion constructs was precipitated on to gold particles and then bombarded on to the onion epidermal cells. The samples were incubated at 23 °C for 24 h in the dark after bombardment. GFP fluorescence was observed and captured using a confocal microscope (Olympus).
GST pull-down assays
Matrix-bound GST-fusion proteins (GST–AtPRMT1a and GST–AtPRMT1b) were incubated with E. coli extracts containing either MBP–AtFib2 or MBP–AtPRMT1b at 4 °C for 2 h with constant gentle mixing. The mixtures were centrifuged at 10000 g for 1 min, and the pellet was washed extensively with modified RIPA buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P40 and 0.5% sodium deoxycholate, with 1 mM PMSF and 0.5 mM dithiothreitol added immediately before use). After the final wash, the pellet was resuspended in an equal volume of 2× SDS loading buffer and separated by SDS/10% PAGE. Proteins were transferred on to a nitrocellulose membrane (Amersham Biosciences) and were immunodetected by monoclonal antibodies against MBP.
RESULTS
Identification of PRMT1 homologues in A. thaliana
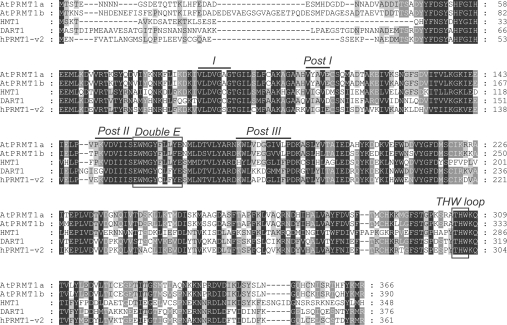
Using BLAST searches, with the amino acid sequence of the hPRMT1 variant 2 (PRMT1-v2) as a query, we identified two proteins, AtPRMT1a and AtPRMT1b, with the closest sequence homology with human PRMT1-v2. These two proteins bear 58 and 56% sequence identity with human PRMT1-v2 respectively, and share 78% identity with and 87% similarity to each other. Amino acid sequences of PRMT1s from Arabidopsis (AtPRMT1a and AtPRMT1b), yeast (Saccharomyces cerevisiae HMT1), Drosophila [DART1 (Drosophila arginine methyltransferase 1)] and human (hPRMT1) were aligned for comparison (Figure 1). Strong conservation was observed particularly within the characteristic SAM-dependent methyltransferase motifs I, post-I, II and III, as well as the ‘double E’ loop and THW (Thr-His-Trp) loop throughout the PRMT family. Major variations were presented in the N-terminus, which may reflect their unique substrate specificities.
Figure 1. Amino acid sequence alignment of PRMT1 from Arabidopsis (AtPRMT1a and AtPRMT1b), yeast (HMT1), Drosophila (DART1) and human (hPRMT1-v2).
Identical amino acids are boxed in black and similar amino acids are boxed in grey. The signature protein methyltransferase motifs I, post I, post II and post III are overlined. The conserved double E loop and THW loop are also boxed. The GenBank® accession numbers used for alignment are: NP_179557 for AtPRMT1a; NP_194680 for AtPRMT1b; CAA84976 for HMT1; NP_650017 for DART1; AAF62893 for hPRMT1-v2.
AtPRMT1a and AtPRMT1b have PRMT activities and specifically methylate histone H4 at R3
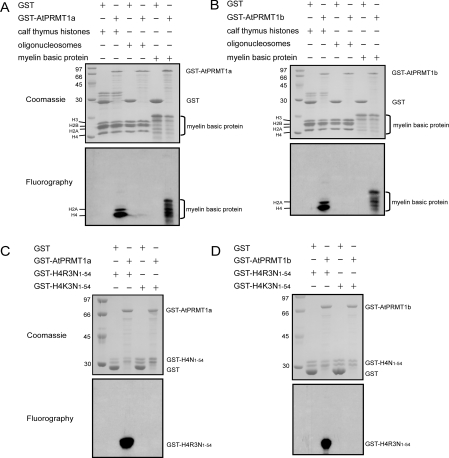
Results from bioinformatics analysis prompted us to examine whether AtPRMT1a and AtPRMT1b contain intrinsic PRMT activities. GST–AtPRMT1a and GST–AtPRMT1b fusion proteins were expressed in E. coli and were affinity-purified using glutathione–Sepharose beads. GST was affinity-purified as a negative control. The matrix-bound GST, and GST–AtPRMT1a and GST–AtPRMT1b fusion proteins were then subjected to a methyltransferase assay in the presence of the methyl donor [3H]SAM and the indicated substrates: core histones from calf thymus, oligonucleosomes purified from HeLa cells and myelin basic protein, which was widely used as substrate in arginine methyltransferase assays. After enzymatic reactions, the mixtures were separated by SDS/PAGE (Figures 2A and 2B, upper panels) and visualized by fluorography (Figures 2A and 2B, lower panels). Histone H4 and myelin basic protein were radioactively labelled by GST–AtPRMT1a and GST–AtPRMT1b only, but not by the GST control. In addition, histone H2A was also methylated, albeit to a lower extent compared with histone H4, as reported previously [8] (Figures 2A and 2B, lower panels). Oligonucleosomes were not methylated in this assay, indicating that other cofactors or a proper conformation may be necessary.
Figure 2. Methyltransferase activities and site specificity of AtPRMT1a and AtPRMT1b in vitro.
Methyltransferase activities of AtPRMT1a (A) and AtPRMT1b (B). GST, GST–AtPRMT1a or GST–AtPRMT1b was immobilized on glutathione–Sepharose beads and incubated with indicated substrates: calf thymus histones, oligonucleosomes and myelin basic protein in the presence of [3H]SAM for 1 h in a final volume of 30 μl of HMT buffer (20 mM Tris/HCl, 4 mM sodium EDTA, 1 mM PMSF and 1 mM dithiothreitol). Methylated proteins were separated by SDS/15% PAGE and stained with Coomassie Blue (A and B, upper panels) and visualized by fluorography (A and B, lower panels). The corresponding proteins are indicated alongside the panels, and molecular masses are indicated in kDa. Both AtPRMT1a (C) and AtPRMT1b (D) specifically methylate histone H4 at R3. Two forms of recombinant histone H4 (GST–H4R3N1–54 and GST–H4K3N1–54) were generated as substrates for AtPRMT1a and AtPRMT1b, and were subjected to the arginine methylation assay as described in the Experimental section. After the reaction, the proteins were separated by SDS/15% PAGE and stained by Coomassie Blue (C and D, upper panels) and visualized by fluorography (C and D, lower panels). The corresponding proteins are indicated on the right of each panel, and molecular masses are indicated on the left in kDa.
Next, we sought to map which sites were subjected to methylation by AtPRMT1a and AtPRMT1b. There are nine arginine residues in the N-terminus of histone H4 (amino acids 1–54, H4N1–54) and R3 is, so far, the only residue on histone H4 reported to be methylated by PRMT1. In this regard, two forms of recombinant substrates were generated: GST fused to the N-terminal peptide covering amino acids 1–54 of histone H4 (GST–H4R3N1–54) and its derivative bearing a mutation at the third arginine to lysine (GST–H4K3N1–54) respectively. Recombinant GST–AtPRMT1a and GST–AtPRMT1b were then incubated with GST–H4R3N1–54 and the mutated form of GST–H4K3N1–54 in the presence of [3H]SAM. Fluorography results showed that GST–H4R3N1–54 was methylated by GST–AtPRMT1a (Figure 2C) and GST–AtPRMT1b (Figure 2D), whereas GST–H4K3N1–54 completely lost its ability to be methylated (Figures 2C and 2D). This result indicated that R3 is the only arginine residue in the N-terminus of histone H4 methylated by AtPRMT1a and AtPRMT1b in vitro.
Methylation on H4-R3 occurs in vivo
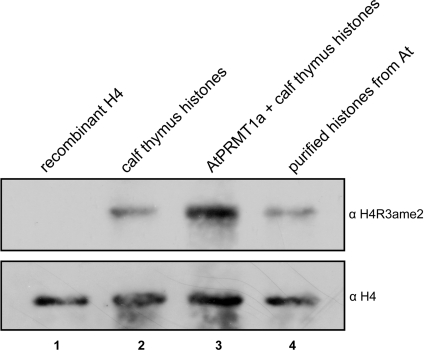
To determine whether endogenous histone H4 is methylated at R3, total histones from Arabidopsis were isolated, and the methylation status of H4-R3 was analysed by Western blotting using anti-(asymmetric dimethyl-H4R3) (anti-H4R3ame2) antibodies. The antibody recognized histone H4 isolated from Arabidopsis well (Figure 3, lane 4, upper panel), but did not recognize recombinant histone H4 expressed in E. coli, which was free from any post-translational modifications (Figure 3, lane 1, upper panel), demonstrating the in vivo occurrence of H4-R3 methylation. As positive controls, calf thymus histone H4 (Figure 3, lane 2, upper panel) and AtPRMT1a-methylated calf thymus histone H4 (Figure 3, lane 3, upper panel) were used, and their levels of methylation were also captured by the anti-H4R3ame2 antibody. The same membrane was re-probed with anti-H4 antibody to demonstrate equal levels of loading (Figure 3, lower panel).
Figure 3. Methylation on H4-R3 occurs in vivo.
Recombinant H4 (upper panel, lane 1) and equivalent amount of calf thymus histones that were either subjected to mock (upper panel, lane 2) or AtPRMT1a methylation (upper panel, lane 3), together with total histones purified from A. thaliana (At) (upper panel, lane 4), were separated by SDS/15% PAGE gel and followed by Western blot analysis using the anti-H4R3ame2 antibody (upper panel). Then the membrane was re-probed with anti-H4 antibody for roughly equal loading (lower panel). All of the relevant proteins are indicated at the top.
AtPRMT1a and AtPRMT1b localize to the nucleus and cytoplasm
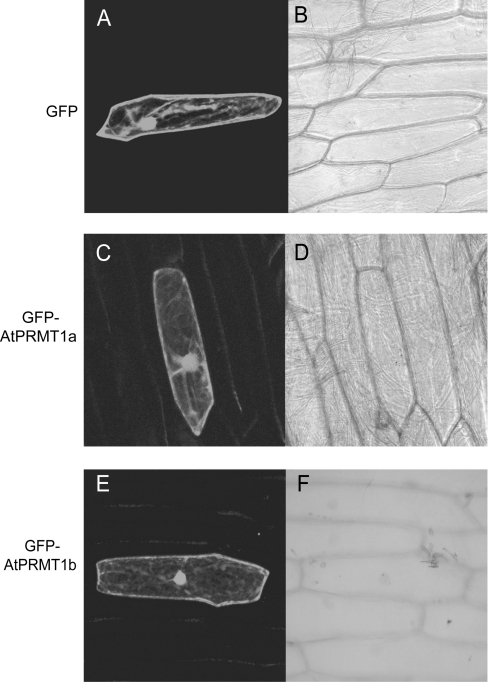
The substrates of AtPRMT1a and AtPRMT1b varied from nuclear proteins (e.g. histones) to cellular membrane proteins (e.g. myelin basic protein), indicating that AtPRMT1a and AtPRMT1b may function in the both nucleus and cytoplasm. To test this, AtPRMT1a and AtPRMT1b cDNAs were fused in-frame to the 5′-end of GFP under the control of the 35S promoter, and the distribution of these fusion proteins in plant cells was determined by a transient transfection system using onion epidermal cells with particle bombardment. As observed by confocal fluorescence microscopy, GFP-fusion proteins of AtPRMT1a and AtPRMT1b displayed a strong nuclear localization, although lacking the nuclear localization signal. As we had expected, both proteins were also dispersed in the cytoplasm (Figures 4C and 4E), similarly to the pattern observed in onion cells expressing GFP alone (Figure 4A).This result was compatible with recent findings from hPRMT1 [34], suggesting that AtPRMT1a and AtPRMT1b may be involved in multiple cellular processes.
Figure 4. AtPRMT1a and AtPRMT1b localize to the nucleus as well as the cytoplasm.
Both AtPRMT1a and AtPRMT1b were fused to the C-terminal end of GFP and transiently transfected into onion epidermal cells. Expression of GFP (A, B), GFP–AtPRMT1a (C, D) and GFP–AtPRMT1b (E, F) were observed by confocal fluorescence microscopy (A, C and E) and brightfield microscopy (B, D and F).
AtPRMT1a interacts with AtPRMT1b in vitro
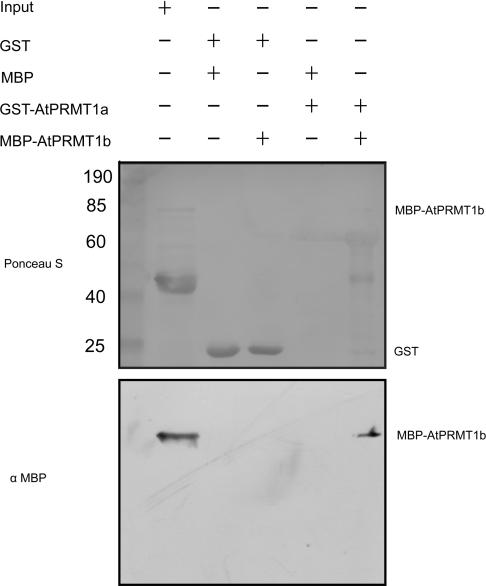
The similar biochemical properties and subcellular distribution between AtPRMT1a and AtPRMT1b suggested that both proteins may be involved in similar physiological progresses. Starting from this premise, a GST pull-down experiment was performed using GST–AtPRMT1a as the ‘bait’ to pull-down MBP–AtPRMT1b. A direct interaction was demonstrated between AtPRMT1a and AtPRMT1b, as no other signal was detected by an anti-MBP monoclonal antibody in any of the control lanes (Figure 5). It was reported that dimerization of hPRMT1 was essential for SAM binding and enzymatic activity, and the dimers further formed an extended oligomer as validated by gel-filtration chromatography [8,35]. In this regard, our result suggested that hetero-oligomers consisting of AtPRMT1a and AtPRMT1b might be formed as the functional unit in regulating plant development.
Figure 5. AtPRMT1a interacts with AtPRMT1b in vitro.
Recombinant GST–AtPRMT1a and MBP–AtPRMT1b were expressed and extracted from E. coli respectively. A GST pull-down assay was performed as described in the Experimental section. The bait–prey protein pairs used in the GST pull-down assay are indicated at the top. GST and MBP alone were used as negative controls. A 10 μl aliquot of E. coli cell extract containing MBP–AtPRMT1b fusion protein (10% input) was used. Upper panel: Ponceau S staining of the nitrocellulose membrane demonstrates the amounts of proteins that were transferred. Molecular masses are indicated in kDa. Lower panel: Western blotting using an anti-MBP monoclonal antibody revealed the interaction between AtPRMT1a and AtPRMT1b.
Both AtPRMT1a and AtPRMT1b asymmetrically methylate and interact with RNA methyltransferase AtFib2
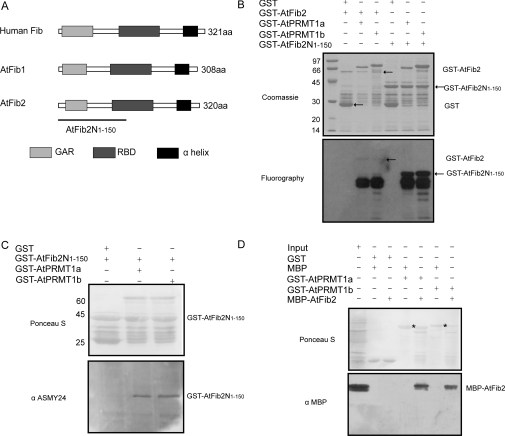
A previous study has revealed components of the ribonucleoprotein complex such as the human RNA methyltransferase Fib (fibrillarin) as the in vitro substrate of PRMT1 [36]. In the genome of Arabidopsis, there are two genes that encode nearly identical proteins, AtFib1 and AtFib2, which function as homologues with human Fib [37]. Both AtFib1 and AtFib2 share the conserved GAR (glycine- and arginine-rich) domain, RBD (RNA-binding domain) and the α-helix domain in analogy to their human counterpart (Figure 6A). To determine whether AtFib2 was a substrate of AtPRMT1a and AtPRMT1b, AtFib2 and its N-terminal region containing the GAR domain (AtFib2N1–150, corresponding to amino acids 1–150 of AtFib2) were cloned and expressed as GST-fusion proteins in E. coli. Purified GST–AtFib2 and GST–AtFib2N1–150 were incubated with GST–AtPRMT1a or GST–AtPRMT1b in the presence of [3H]SAM as previously described. The reaction products were separated by SDS/PAGE and visualized by fluorography. As shown in Figure 6(B), both AtFib2 and its N-terminal region AtFib2N1–150 were methylated by AtPRMT1a and AtPRMT1b in vitro, indicating that AtFib2 may be the potential substrate of AtPRMT1. Since GST–AtFib2 fusion protein (approx. 60 kDa) is very sensitive to proteolysis during expression and purification, and is less competitive to acquire [3H]SAM, the level of methylation of full-length GST–AtFib2 is minimal, the major methylated product (degraded fragments of GST–AtFib2) electrophorese as a broad band ranging from 28 to 36 kDa, as reported previously [38]. Other groups' work has also demonstrated the GST–GAR fusion protein or fusion protein containing the GAR domain such as GST–rpS2 (ribosomal protein S2) are very susceptible to degradation [39,40]. These fragments are much more competitive to acquire [3H]SAM than full-length GST–AtFib2, which ultimately results in the minimal level of methylation of full-length GST–AtFib2 (Figure 6B).
Figure 6. AtPRMT1a and AtPRMT1b methylate and interact with AtFib2.
(A) Schematic diagram of the domain structures of human Fib, AtFib1 and AtFib2. The conserved GAR domains are shown as light grey boxes, the RBDs are shown as dark grey boxes, and the α-helix domains are shown as black boxes. The N-terminal region of AtFib2 corresponding to amino acids 1–150 (AtFib2N1–150) used as the substrate in the methyltransferase assay is indicated by the line. (B) GST–AtFib2 and GST–AtFib2N1–150 were incubated with GST–AtPRMT1a and GST–AtPRMT1b separately in the presence of [3H]SAM. Methylated proteins were separated by SDS/10% PAGE (upper panel) and visualized by fluorography (lower panel). The positions of corresponding proteins are indicated on the right of each panel. Molecular masses are indicated on the left in kDa. (C) AtPRMT1a and AtPRMT1b are Type I PRMTs. Ponceau S staining of the nitrocellulose membrane demonstrated roughly equal loading (upper panel). ASYM24 antibody against asymmetrically dimethylated arginine residues recognized GST–AtFib2N1–150 methylated by AtPRMT1a and AtPRMT1b, not by GST control (lower panel). Molecular masses are indicated in kDa. (D) Ponceau S staining of the nitrocellulose membrane after GST pull-down assays using GST–AtPRMT1a and GST–AtPRMT1b as bait and MBP–AtFib2 as prey (upper panel). The bait–prey protein pairs used in the GST pull-down assay are indicated at the top. GST–AtPRMT1a and GST–AtPRMT1b are indicated by an asterisk. A 10 μl aliquot of E. coli cell extract containing MBP–AtFib2 fusion protein (10% input) was used. The membrane was probed with an anti-MBP monoclonal antibody (lower panel).
To determine the type of activity that AtPRMT1a and AtPRMT1b catalyse, GST–AtFib2N1–150 was subjected to Western blotting after a methyltransferase activity assay: the samples were separated by SDS/15% PAGE and transferred on to nitrocellulose membrane and probed with an antibody specific for asymmetrically dimethylated arginine residues (ASYM24, Upstate). The result demonstrated the presence of asymmetric dimethylation on GST–AtFib2N1–150, suggesting AtPRMT1a and AtPRMT1b are Type I arginine methyltransferases (Figure 6C).
Since human Fib was reported to associate with a subcomplex containing PRMT5, PRMT1, SF2A-p32 (splicing factor SF2-associated protein p32), tubulin α3 and tubulin β1 [41], we determined to assess whether there existed a direct interaction between AtFib2 and AtPRMT1a/AtPRMT1b. To this end, GST–AtPRMT1a and GST–AtPRMT1b proteins were immobilized on glutathione–Sepharose beads as the ‘bait’, and the MBP–AtFib2 was expressed as the ‘prey’ in GST pull-down assays. As detected by Western blotting using monoclonal antibodies against the MBP tag, MBP–AtFib2 was shown to be captured by GST–AtPRMT1a or GST–AtPRMT1b, but not by the GST control (Figure 6D), indicating that AtFib2 interacted directly with GST–AtPRMT1a and GST–AtPRMT1b in vitro.
DISCUSSION
In the present study, we have identified two genes encoding PRMTs, AtPRMT1a and AtPRMT1b, in Arabidopsis, both of which exhibit the highest homology with hPRMT1. Phylogenetic analysis of PRMT1s from different species shows that these proteins are highly conserved from yeasts to plants and animals with respect to amino acid sequences and possibly their functions: they all share the signature SAM-dependent methyltransferase motifs I, post-I, II, III and the conserved double E loop as well as the THW loop, and subsequently demonstrate methyltransferase activities when assayed in vitro. The least conserved region among PRMT1s is the N-terminus.
Our biochemical evidence showed that both AtPRMT1a and AtPRMT1b belonged to the Type I PRMT and specifically methylated R3 on histone H4, since mutation of this residue to lysine completely abolished the ability of AtPRMT1a and AtPRMT1b to methylate H4 in vitro. Although the in vitro histone methyltransferase assays detected no activity using oligonucleosomes, the basic unit of chromatin as substrate, Western blotting using an antibody directed against H4-R3 does demonstrate the in vivo occurrence of methylation on this site, suggesting that other cofactor(s) or proper structural conformation of the chromatin may be necessary for AtPRMT1a and AtPRMT1b to execute enzymatic activities in vivo. Further investigation concerning global changes in H4-R3 methylation levels of histones extracted from AtPRMT1a/AtPRMT1b double mutants or overexpressing transgenic plants would promisingly uncover the specific catalytic property of AtPRMT1a and AtPRMT1b at this site.
Methylation on histone H4-R3 by PRMT1 has been regarded as a conserved post-translational modification throughout eukary-otes [42]. Besides AtPRMT1a and AtPRMT1b reported in the present paper, there is another arginine methyltransferase that is responsible for the methylation on H4-R3 in Arabidopsis, which is designated as AtPRMT5/SKB1. Notably, AtPRMT5/SKB1-mediated symmetric dimethylation on H4-R3 controls flowering time by repressing the transcription of FLC [30,31]. Although, in plants, the biological consequence of asymmetric methylation on H4-R3 remains to be elucidated, studies in mammals have provided clues as to the correlation between H4-R3 methylation and transcriptional activation [8,15,20]. Given the high conservation in eukaryotic organisms, it is reasonable to speculate that AtPRMT1a and AtPRMT1b may also play essential roles in activating gene transcription. If that is the case, it would be very interesting, since methylation at the same site (H4-R3) brings the opposite biological consequences.
In addition to histone H4, our methylation assays have also revealed histone H2A as a substrate for AtPRMT1a and AtPRMT1b, as reported previously [8]. Histone H2A undergoes post-translational modifications such as phosphorylation and ubiquitination. Phosphorylation of histone H2A has been suggested to correlate with DNA repair [43] and cell-cycle progression [44], whereas ubiquitination of H2A has been demonstrated to play a role in polycomb silencing [45] and X chromosome inactivation [46]. Although a methylated signal for endogenous histone H2A has not yet been detected, interestingly, H2A harbours the same extreme N-terminus sequence SGRGK as that of H4 and may be also methylated at R3 since methylated H2A could also be recognized by anti-H4R3ame2 antibody [8]. To date, the precise function of histone H2A methylation remains to be clarified, thus obtaining more evidence to support the in vivo existence of arginine methylation on histone H2A and mapping the sites that are subjected to methylation may hopefully address its biological outputs in plants.
Gene duplication is quite a common event during evolution [47]. After gene duplication, divergence may take place in various ways: (i) only one copy functions normally, while the other turns into a pseudogene and loses its function; (ii) both duplicated genes are closely linked and are very similar in function; or (iii) each daughter gene evolves its new functions at different tissues and developmental stages [48,49]. Since two copies of AtPRMT1 (AtPRMT1a on chromosome 2 and AtPRMT1b on chromosome 4) were identified in the Arabidopsis genome with nearly identical biochemical properties, we analysed the subcellular distribution of these two proteins and revealed that both of them localized to the nucleus as well as to the cytoplasm. This result is consistent with that observed in hPRMT1, indicating that AtPRMT1a and AtPTMT1b may also participate in multiple physiological events by catalysing the methylation of a variety of proteins inside and outside the nucleus. In addition, our GST pull-down experiment demonstrated that AtPRMT1a interacted with AtPRMT1b. Previously, yeast HMT1 and hPRMT1 were reported to exist as hexamers [50] and homodimers or higher-order homo-oligomers [8,35] respectively. Given the fact that dimerization is essential for methyltransferase activity [51], we propose these two proteins might potentially form homodimers and further form into hetero-oligomers as the functional unit, so that the physical interaction can be detected in GST pull-down assays. Further investigations to monitor the expression pattern of AtPRMT1a and AtPRMT1b genes and co-localization of the two proteins might provide much more evidence with regard to whether the hetero-oligomers represent the functional unit in the process of plant development, even though it is not yet clear whether there are any differences in the substrate specificities between AtPRMT1a and AtPRMT1b.
Like their human counterpart hPRMT1, both AtPRMT1a and AtPRMT1b methylate not only the histones, but also other proteins, such as the RNA methyltransferase AtFib2 in vitro. Fib is highly conserved from yeasts to plants and animals that are associated with box C/D snoRNPs (small nucleolar ribonucleoproteins) and directs pre-rRNA 2′-O-ribose methylation, processing and ultimately ribosome assembly [37,52]. Furthermore, our GST pull-down assay revealed that AtPRMT1a and AtPRMT1b interact with AtFib2 in vitro. This result not only supports the previous finding that human Fib associated with PRMT1 and PRMT5 in a subcomplex [38], but also demonstrates, for the first time, a direct linkage between the PRMT (AtPRMT1a/AtPRMT1b) and the RNA methyltransferase (AtFib2) in plants. It is intriguing to dissect whether and how AtPRMT1a and AtPRMT1b affect AtFib2 function via methylating its GAR domain, and how protein methyltransferases collaborate with RNA methyltransferase in the processes of snoRNP assembly and ribosome biogenesis.
Acknowledgments
We thank Bin Liu, Chunyan Liu, Yong Ding and Falong Lu in the laboratory of X. C. for comments on this manuscript. We are also grateful to Dr Dongqiao Shi from Dr Weicai Yang's laboratory of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, and Dr Xia Wang at the Institute of Genetics and Developmental Biology for help in confocal microscopy. The oligonucleosomes and the pAVA321GFP vector were gifts from Dr Yi Zhang (University of North Carolina at Chapel Hill, NC, U.S.A.) and Dr Xuemei Chen (University of California at Riverside, CA, U.S.A.) respectively. This research was supported by the National Basic Research Program of China (grant number 2005CB522400) and National Natural Science Foundation of China (grant numbers 30430410, 30325015 and 30621001) to X. C. and the Chinese Academy of Sciences (grant number CXTD-S2005-2) to X. C.
References
- 1.Shiio Y., Eisenman R. N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 4.Huang S., Litt M., Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedkov Y., Cho E., Petruk S., Cherbas L., Smith S. T., Jones R. S., Cherbas P., Canaani E., Jaynes J. B., Mazo A. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang J., Feng Q., Ketel C. S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J. A., Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 7.Tamaru H., Selker E. U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 9.Bedford M. T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Yu M. C., Lamming D. W., Eskin J. A., Sinclair D. A., Silver P. A. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 2006;20:3249–3254. doi: 10.1101/gad.1495206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres-Padilla M. E., Parfitt D. E., Kouzarides T., Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gary J. D., Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 13.Pollack B. P., Kotenko S. V., He W., Izotova L. S., Barnoski B. L., Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 14.Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An W., Kim J., Roeder R. G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J., Kao P. N., Herschman H. R. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 2000;275:19866–19876. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- 18.Lin W. J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 19.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M. R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrero M. J., Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawlak M. R., Scherer C. A., Chen J., Roshon M. J., Ruley H. E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 2000;20:4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mowen K. A., Schurter B. T., Fathman J. W., David M., Glimcher L. H. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell. 2004;15:559–571. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 23.Xu W., Cho H., Kadam S., Banayo E. M., Anderson S., Yates J. R., 3rd, Emerson B. M., Evans R. M. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cote J., Boisvert F. M., Boulanger M. C., Bedford M. T., Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell. 2003;14:274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith W. A., Schurter B. T., Wong-Staal F., David M. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem. 2004;279:22795–22798. doi: 10.1074/jbc.C300512200. [DOI] [PubMed] [Google Scholar]
- 26.Shen E. C., Henry M. F., Weiss V. H., Valentini S. R., Silver P. A., Lee M. S. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boisvert F. M., Dery U., Masson J. Y., Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 29.Yu M. C., Bachand F., McBride A. E., Komili S., Casolari J. M., Silver P. A. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei Y., Niu L., Lu F., Liu C., Zhai J., Kong X., Cao X. Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., Liu C., Bi Y., Qian Q., Cheng Z., Chu C., Cao X. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007;19:9–22. doi: 10.1105/tpc.106.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumbusch L. O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R. B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrmann F., Lee J., Bedford M. T., Fackelmayer F. O. Dynamics of human protein arginine methyltransferase 1 (PRMT1) in vivo. J. Biol. Chem. 2005;280:38005–38010. doi: 10.1074/jbc.M502458200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X., Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C. H., Huang H. M., Hsieh M., Pollard K. M., Li C. Arginine methylation of recombinant murine fibrillarin by protein arginine methyltransferase. J. Protein Chem. 2002;21:447–453. doi: 10.1023/a:1021394903025. [DOI] [PubMed] [Google Scholar]
- 37.Barneche F., Steinmetz F., Echeverria M. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 2000;275:27212–27220. doi: 10.1074/jbc.M002996200. [DOI] [PubMed] [Google Scholar]
- 38.Tang J., Gary J. D., Clarke S., Herschman H. R. PRMT 3, a Type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 39.Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 40.Swiercz R., Person M. D., Bedford M. T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem. J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanagida M., Hayano T., Yamauchi Y., Shinkawa T., Natsume T., Isobe T., Takahashi N. Human fibrillarin forms a sub-complex with splicing factor 2-associated p32, protein arginine methyltransferases, and tubulins α3 and β1 that is independent of its association with preribosomal ribonucleoprotein complexes. J. Biol. Chem. 2004;279:1607–1614. doi: 10.1074/jbc.M305604200. [DOI] [PubMed] [Google Scholar]
- 42.Strahl B. D., Briggs S. D., Brame C. J., Caldwell J. A., Koh S. S., Ma H., Cook R. G., Shabanowitz J., Hunt D. F., Stallcup M. R., Allis C. D. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 43.Downs J. A., Lowndes N. F., Jackson S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 44.Aihara H., Nakagawa T., Yasui K., Ohta T., Hirose S., Dhomae N., Takio K., Kaneko M., Takeshima Y., Muramatsu M., Ito T. Nucleosomal histone kinase-1 phosphorylates H2A Thr119 during mitosis in the early Drosophila embryo. Genes Dev. 2004;18:877–888. doi: 10.1101/gad.1184604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 46.Fang J., Chen T., Chadwick B., Li E., Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 47.Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin. Cell Dev. Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- 48.Hughes A. L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 49.Raes J., van de Peer Y. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinformatics. 2003;2:91–101. [PubMed] [Google Scholar]
- 50.Weiss V. H., McBride A. E., Soriano M. A., Filman D. J., Silver P. A., Hogle J. M. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 51.Lee D. Y., Ianculescu I., Purcell D., Zhang X., Cheng X., Stallcup M. R. Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function. Mol. Endocrinol. 2007;21:1381–1393. doi: 10.1210/me.2006-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]