Abstract
Lipocalins are extracellular proteins (17–25 kDa) that bind and transport small lipophilic molecules. The three-dimensional structure of the first lipocalin from a metatherian has been determined at different values of pH both with and without bound ligands. Trichosurin, a protein from the milk whey of the common brushtail possum, Trichosurus vulpecula, has been recombinantly expressed in Escherichia coli, refolded from inclusion bodies, purified and crystallized at two different pH values. The three-dimensional structure of trichosurin was solved by X-ray crystallography in two different crystal forms to 1.9 Å (1 Å=0.1 nm) and 2.6 Å resolution, from crystals grown at low and high pH values respectively. Trichosurin has the typical lipocalin fold, an eight-stranded anti-parallel β-barrel but dimerizes in an orientation that has not been seen previously. The putative binding pocket in the centre of the β-barrel is well-defined in both high and low pH structures and is occupied by water molecules along with isopropanol molecules from the crystallization medium. Trichosurin was also co-crystallized with a number of small molecule ligands and structures were determined with 2-naphthol and 4-ethylphenol bound in the centre of the β-barrel. The binding of phenolic compounds by trichosurin provides clues to the function of this important marsupial milk protein, which is highly conserved across metatherians.
Keywords: fluorescence, lipocalin, metatherian lactation, opossum, protein structure, X-ray crystallography
Abbreviations: BLG, β-lactoglobulin; MUP, major urinary protein; NGAL, neutrophil gelatinase-associated lipocalin; rmsd, root mean square deviation
INTRODUCTION
The metatherians have historically been regarded by science as curiosities, the name quite literally means ‘other beasts’. Among the metatherians, the biology of the opossum has recently come to the fore as it occupies a unique position on the evolutionary tree, sharing a common ancestor with humans approx. 130 million years ago. This is the midpoint between the divergence of placental mammals such as humans and mice which have a common ancestor approx. 80–90 million years ago, and non-mammalian animals which diverged from humans 300–350 million years ago. The unique biology of the early development of opossum young (and other metatherians) also provides impetus for research. Research into the milk proteins of Trichosurus vulpecula, the common brushtail possum, stems from a concerted effort to study the physiological differences between metatherians and other mammals. T. vulpecula, has been widely studied in both Australia and New Zealand from both an academic evolutionary point of view as well as an applied environmental perspective. It was introduced to New Zealand from its native Australia in the middle of the 19th century to establish a fur trade [1]. It has since become widely established, damaging large areas of forest every year and exerting considerable resource pressure upon New Zealand's endemic fauna [2–4]. Analogous to the badger in the U.K., the possum in New Zealand also acts as an efficient vector for bovine tuberculosis. These factors make the brushtail possum one of the most important vertebrate pests in New Zealand.
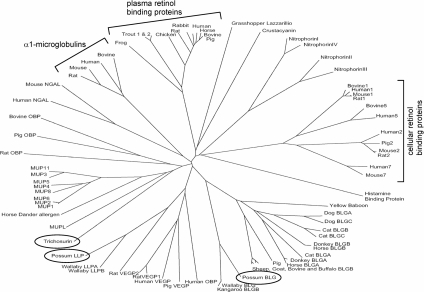
Marsupials have invested in lactation, as opposed to placentation, as a way of rearing their young. Reproduction in the Australian brushtail possum (T. vulpecula) is typified by a short gestation (17 days) followed by a prolonged period of lactation (>200 days) [5]. Lactation is divided into at least three distinct phases between which milk composition changes dramatically [1,6,7]. Possum milk contains a large number of whey proteins, most of which are homologues of eutherian milk proteins [8–10]. Trichosurin is one of three predominant lipocalins found in the milk of T. vulpecula [10] (Figure 1). It bears significant similarity (34% amino acid identity) to the rodent MUPs (major urinary proteins) and to the major horse dander allergen equ_c1 (28% amino acid identity). More recently, trichosurin homologues have been identified in both the tammar wallaby (87% amino acid identity) and the South American opossum (77% amino acid identity). This high level of conservation in species that diverged approx. 80 million years ago due to continental separation suggests that the function of trichosurin is critical in metatherian lactation. Currently, its biochemical and biological functions are not understood.
Figure 1. Sequence tree showing relationships among the lipocalin superfamily.
Sequence tree showing relationships among 74 animal lipocalins (possum milk lipocalins are circled). BLGA/BLGB; β-lactoglobulins A or B; LLPA/LLPB; late lactation protein A or B; OBP; odourant-binding protein, VEGP; von Ebners gland protein. Figure prepared using ClustalX [43].
The lipocalins are a large family of proteins (diverse in sequence but structurally homologous) that bind and transport small hydrophobic molecules in a central hydrophobic pocket [11,12]. The binding specificity exhibited by lipocalins varies widely across the family. For example, BLG (β-lactoglobulin) is known to bind a wide array of molecules from retinal to fatty acids [13], whereas other lipocalins such as the MUPs from mice and rats bind a smaller range of compounds in a manner more in keeping with their role in scent-based territory marking [14]. The internal topology of the hydrophobic pocket is a key factor in the type and selectivity of binding, as is access to the binding pocket which is usually controlled by residues forming a loop between β-strands at one end of the lipocalin β-barrel.
The lipocalin superfamily is defined by a common fold, an eight-stranded anti-parallel β-barrel, with variable loop regions flanking and enclosing the top and bottom of the barrel to form a hydrophobic pocket in the interior [15]. A conserved disulfide bond links one of the C-terminal β-strands with the C-terminus. Identification of lipocalins is generally made via the presence of three small conserved sequence motifs which are also used to demarcate family membership between ‘core’ lipocalins and ‘outliers’, based on whether all three, or just the first two, sequence motifs are present.
In the present study, we provide the first three-dimensional structure of a metatherian lipocalin, trichosurin. This structure was solved by X-ray crystallography in two different crystal forms at two disparate values of pH (pH=4.6 and pH=8.2). To investigate the binding of ligands by trichosurin, six small molecules known to bind within the barrel of the closely related rodent MUPs have been used as molecular probes. Two of these small molecules were shown by fluorescence to bind to trichosurin, and we present the three-dimensional structures of trichosurin with 2-naphthol and 4-ethylphenol bound in the central pocket of the β-barrel. 2-Naphthol binds in the cavity of trichosurin with micromolar affinity and a preference for small phenolic compounds suggests a possible biological role for trichosurin.
EXPERIMENTAL
Protein expression and refolding
DNA encoding the mature trichosurin protein was amplified by PCR from a possum cDNA library and cloned into the vector pGEMTeasy. The trichosurin gene was then subcloned into the expression vector pET23a which was transformed into E. coli cells [BL21 (DE3)] carrying the pRI rare codon plasmid. Small overnight cultures (5 ml) were used to inoculate 1 litre cultures and these were grown at 37 °C and protein expression was subsequently induced at an D590 of 0.5–0.7 using 1 mM IPTG (isopropyl β-D-thiogalactoside). At 3 h post-induction, cells were harvested by centrifugation (6000 g at 4 °C for 10 min) and cell pellets were washed with 50 mM Tris/HCl (pH 8.2) and 50 mM NaCl, then repelleted and frozen at −20 °C. Frozen cell pellets were thawed and re-suspended in buffer (50 mM Tris/HCl, pH 8.2, and 50 mM NaCl), then lysed by passing once through a Constant Systems cell disruptor at 25 kPa. The cell lysate was then centrifuged at 10000 g for 10 min to pellet the insoluble inclusion bodies. The inclusion bodies were washed by resuspending them in buffered 0.1% (v/v) Triton X-100, incubated on ice for 10 min, centrifuged (10000 g at 4 °C for 10 min) and then suspensed again in buffer. This was then treated with 50 ml of 8 M urea and 50 mM Tris/HCl (pH 8.2), and incubated for 1 h on ice to denature and solubilize the inclusion bodies. The denatured protein was refolded by dialysis at 18 °C against 1 litre of 50 mM Tris/HCl (pH 8.2), 50 mM NaCl, 100 mM sucrose, 1 mM EDTA, 1 mM GSH and 0.1 mM GSSG. After 8 h the buffer was changed to 1 litre of 50 mM Tris/HCl (pH 8.2) and 50 mM NaCl.
Protein purification
The refolded trichosurin was bound to a HiTrap Q column (Pharmacia Biotech) and then eluted using a gradient of 50–500 mM NaCl. Fractions containing trichosurin were pooled and concentrated. The protein was then purified in a second step by gel-filtration chromatography (Superdex75 16/60; Pharmcia Biotech) in 50 mM Tris/HCl (pH 8.2) and 150 mM NaCl. The protein quality in each fraction was assessed by dynamic light scattering, and those fractions with the highest degree of monodispersity were pooled and concentrated to ∼10 mg/ml for use in crystallization screening. N-terminal sequencing confirmed that the N-terminus of the protein was intact.
Crystallization
Initial crystallization screening was carried out by the hanging drop vapour diffusion method in 24-well format and using the Hampton I crystallization screen [16]. Drops of 2 μl were set up by mixing 1 μl of trichosurin (10 mg/ml) with 1 μl of reservoir solution. Successful crystallization conditions from the initial screen were refined and diffraction quality crystals were grown from drops consisting of 3 μl of protein/3 μl of reservoir solution [100 mM Tris/HCl (pH 8.2), 200 mM Li2SO4 and 10–14% poly(ethylene glycol) 4000]. Crystallization was also refined at low pH values, with diffraction-quality crystals being obtained using a precipitant solution of 100 mM sodium acetate (pH 4.6), 200 mM CaCl2 and 6–8% (v/v) 2-propanol.
Ligand complexes of trichosurin were obtained by co-crystallization. Each ligand was dissolved in 2-propanol at a concentration of 1 M, and this solution was added to the reservoir solution immediately prior to crystallization in sufficient quantity to give a final ligand concentration in each case of 3–5 mM. Crystallization was at low pH, using the conditions given above, except that ZnCl2 replaced the CaCl2 from the native crystallization condition.
Data collection
For apo-trichosurin crystals, X-ray diffraction data were collected at 113 K using crystals which had first been soaked briefly in cryoprotectant consisting of mother liquor (for crystals grown at pH 4.6 the mother liquor is precipitant solution described above; at pH 8.2 the mother liquor is reservoir solution described above) containing 32% glycerol (for crystals grown at pH 4.6) or mother liquor containing 22% (v/v) glycerol (for crystals grown at pH 8.2). Diffraction images were collected using a MAR345 detector with Cu-Kα X-rays from a Rigaku H3R rotating anode generator. Data for the trichosurin–ligand complex crystals were collected at the ESRF (European Synchrotron Radiation Facility, Grenoble, France). These synchrotron data were collected at 113 K on beamline ID 14-4 using an ADSC Quantum4 Q4R CCD (charge-coupled-device) camera. For data collection statistics see Table 1.
Table 1. Data collection and refinement statistics.
Numbers in parentheses indicate statistics for the outer resolution shell. trich, trichosurin.
| Apo-trich4.6 | Apo-trich8.2 | 2Nap-trich4.6 | 4Eth-trich4.6 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P43212 | P21 | P21 |
| Unit cell (Å) | 57.85, 44.58, 70.41 | 100.16, 100.16, 140.55 | 44.3, 100.4, 83.69 | 44.3, 100.4, 84.9 |
| 90°, 101.5°, 90° | 90°, 90°, 90° | 90°, 90.03°, 90° | 90°, 90.03°, 90° | |
| Wavelength (Å) | 1.54 | 1.54 | 0.89 | 0.89 |
| Resolution (Å) | 25–1.9 (1.97–1.90) | 20–2.5 (2.59–2.50) | 15–1.5 (1.6–1.5) | 30–1.5 (1.6–1.5) |
| Rmerge | 0.043 (0.192) | 0.085 (0.516) | 0.086 (0.321) | 0.072 (0.289) |
| I/σ(I) | 17.6 (6.6) | 33.9 (6.4) | 9.9 (4.9) | 11.6 (4.3) |
| Completeness (%) | 95.1 (88.9) | 100 (100) | 96.9 (88) | 98.6 (96) |
| Nobs | 108260 | 534113 | 284263 | 521056 |
| Nunique | 26645 | 25922 | 92603 | 116743 |
| Refinement | ||||
| Rwork (%) | 21.6 (31.1) | 22.0 (27.3) | 22.6 (30.9) | 22.7(33.3) |
| Rfree (%) | 24.9 (35.3) | 28.6 (36.9) | 24.0 (33.9) | 24.5 (33.9) |
| B (Å2) protein | 28.4 | 33.7 | 24.2 | 22.1 |
| B (Å2) solvent | 33.7 | 32.0 | 33.7 | 32.7 |
| B (Å2) ligand | 23.8 | 34.0 | ||
| Natoms | 2373 | 4883 | 5124 | 5248 |
| Rmsd statistics | ||||
| Bond length (Å) | 0.0052 | 0.0065 | 0.006 | 0.005 |
| Bond angle (°) | 1.26 | 1.20 | 1.24 | 1.20 |
Structure solution, model building and model refinement
Diffraction data were processed using DENZO and scaled using SCALEPACK from the HKL suite [17]. The space group and unit cell dimensions for the data from crystals grown at pH 4.6 were P21, a=57.85 Å (1 Å=0.1 nm), b=44.58 Å, c=70.41 Å, β=101.5°, with two molecules in the asymmetric unit. Similarly, for crystals grown at pH 8.2 the space group and unit cell dimensions were P43212, a=b=101.16 Å, c=140.56 Å, with four molecules in the asymmetric unit.
The structure of apo-trichosurin (crystals grown at pH 4.6) was solved by molecular replacement with AMoRe [18], using the rat α2U-globulin structure (PDB entry: 2A2U) as a search model. Two solutions were found, corresponding to two molecules in the asymmetric unit and a Matthews coefficient (VM) of 2.3 Å3/Da. The two molecules were related by a local 2-fold rotation axis and positioned such that a dimer interface is clearly defined. Initial automated model building was with wARP [19] which built a model comprising 214 of the expected 332 residues (side chains were automatically added by wARP for 210 residues). A further 72 residues were added during iterations of building and refinement using O [20] and CNS [21] respectively. The structure of high-pH apo-trichosurin was solved using the low-pH apo-trichosurin coordinates and molecular replacement. Similarly, the structures of trichosurin with bound substrate were solved by molecular replacement (AMoRe) using the low-pH trichosurin co-ordinates as a search model. In all cases refinement and model-building were carried out in CNS and O [20,21].
The small molecules for ligand-binding experiments were thymol, trans-2-nonenal, 2-naphthol, 4-ethylphenol, 2-isobutyl-3-methoxy pyrazine and 2-sec-butyl thiazole and were purchased from Sigma–Aldrich.
Ligand binding using fluorescence
Fluorescence binding experiments were carried out using a Hitachi F-4500 fluorescence spectrophotometer and a square quartz cuvette with a 5 mm path length. Trichosurin was used at a concentration of 20 μM in 10 mM sodium phosphate buffer (pH 7.96). Stock solutions of substrates were made up to 1 mM in ethanol and added to 400 μl aliquots of protein to a final concentration of 20 μM. Changes in fluorescence maxima were observed using an excitation wavelength of 282 nm by comparison against appropriate reference spectra of trichosurin and trichosurin with 2% ethanol.
RESULTS AND DISCUSSION
Structure determination
Trichosurin was expressed in E. coli as insoluble inclusion bodies, denatured in 8 M urea and refolded by dialysis against a suitable refolding buffer. Washing the inclusion bodies and then refolding had the advantage of producing very pure protein in the absence of an affinity purification tag. Following the final size-exclusion purification step, the protein showed excellent monodispersity as determined by dynamic light scattering.
Molecular replacement using the co-ordinates for α2U-globulin (PDB code: 2A2U) as a search model allowed the structure of trichosurin to be solved using the data collected from a single crystal grown at pH 4.6. This model, termed apo-trich4.6, consisted of residues 24–166 of trichosurin, the first 23 residues were unable to be modelled (these residues are present as shown by N-terminal sequencing prior to crystallization). The model shows good geometry: a Ramachandran plot of chains A and B showed that residues lie mainly in the most favoured regions of the plot with 86% in core and 12.8% in allowable regions. 1.2% of the residues are in generously allowed regions and there are no outliers. The model was refined at 1.9 Å resolution, giving an R-factor of 21.6% (Rfree=24.8%).
The structure of trichosurin crystallized at pH 8.2, termed apo-trich8.2, was solved by molecular replacement using the completed apo-trich4.6 monomer. The rationale for determining the structure of trichosurin at high pH was to look for conformational changes associated with pH changes similar to that seen for bovine β-lactoglobulin, as well as assessing the stability of the dimer under various pH conditions. The structure was refined at 2.5 Å resolution to a final R-factor of 22.0% (Rfree=28.6%). The Ramachandran plot for the apo-trich8.2 model showed 84.3% of the residues were in core regions of the plot, 14.7% in allowable regions and 0.9% in generously allowed regions. Full data collection and model refinement statistics are given in Table 1.
Crystals of the 2-naphthol– and 4-ethylphenol–trichosurin complexes diffracted to 1.5 and 1.6 Å resolution respectively, at the ESRF. The crystals of the two ligand complexes proved to be isomorphous, as were crystals grown in the presence of thymol, 2-isobutyl-3-methoxy pyrazine, 2-sec-butyl thiazole and trans-2-nonenal; both fluorescence measurements and scrutiny of the electron density maps (after co-crystallization) showed that these latter four compounds did not bind to trichosurin.
The structures of the two ligand complexes were solved by molecular replacement using the apo-trich4.6 structure as a search model. The model containing 2-naphthol was refined to a final R-value of 22.6% (Rfree=24.0%) at 1.5 Å resolution, whereas the structure containing 4-ethylphenol as a ligand was refined to a final R-value of 22.7% (Rfree=24.5%) at 1.6 Å resolution.
Molecular structure
Recombinant trichosurin consists of 166 amino acids, including the addition of an N-terminal methionine which is a product of the subcloning and prokaryotic expression. The apo-trich4.6 structure accounts for 143 of these amino acids with the first 23 amino acids being unable to be traced due to sketchy or absent electron density. For apo-trich8.2 the first 20 amino acids of the peptide chain are absent. The two structures are very similar, with an rmsd (root mean square difference) in Cα atom positions of 0.54 Å over residues Leu24 to Cys166.
As expected, the overall structural fold of trichosurin is an eight-stranded, anti-parallel β-barrel with a helix near the C-terminus which is common to all lipocalins (see Figure 2A). An extended loop region between β-strands A and B forms a short helix and an open turn which, together with residues on the loop between β-strands E and F, close one end of the barrel. Residues which are N-terminal to β-strand A pack against residues in the bottom of the barrel, effectively sealing the other end of the barrel. A disulfide bond, the last major conserved feature of the lipocalin fold, is formed between Cys73 and Cys166 and ties the C-terminus to β-strand D.
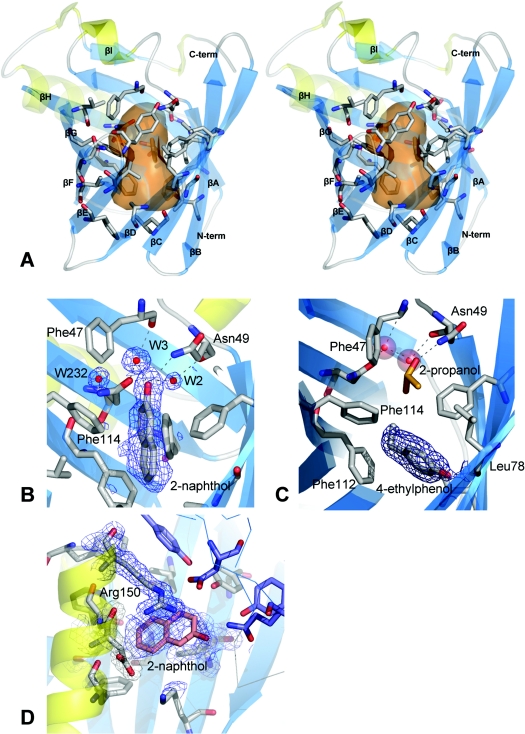
Figure 2. Trichosurin structure, topology and ligand binding.
(A) Stereo diagram showing the trichosurin monomer, with β-strands in blue (labelled βA–H) and α-helices yellow. The residues whose side chains define the binding pocket are rendered as grey sticks with oxygen atoms in red and nitrogen in blue. The transparent orange surface represents the approximate size of the internal cavity as a molecular contact surface described by a sphere of 1.4 Å radius. (B) A 2(Fo)−(Fc) electron density map contoured at 1σ depicted in blue mesh showing the 2-naphthol ligand bound within the trichosurin barrel and the hydrogen bonds made to water W3 and Asn49 ND1. (C) 2(Fo)−(Fc) electron density map contoured at 1σ showing 4-ethylphenol (surrounded by electron density mesh) donating a hydrogen bond to Leu78 O and occupying the lower part of the trichosurin-binding pocket. A 2-propanol molecule (orange and red) is still bound in the upper part of the binding pocket along with the water molecules W2 and W3 (red spheres). (D) The second binding site for 2-naphthol between the helix, the outside of the barrel and the dimer interface. The blue mesh is from a 2(Fo)−(Fc) electron density map contoured at 1σ. The thin blue lines and the purple residues are from the second molecule of the dimer.
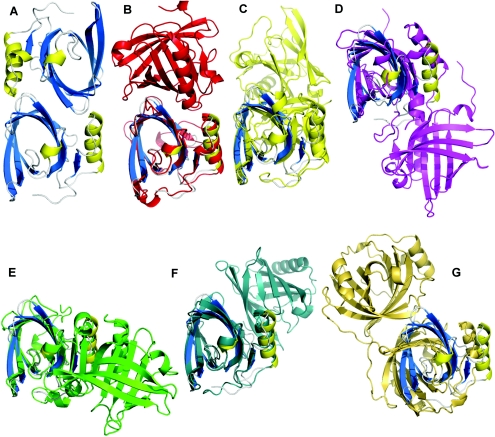
Trichosurin forms an unusual dimer which has not previously been seen among structures from the lipocalin superfamily. The mode of dimerization is conserved between structures determined at pH 4.6 and pH 8.2 and additional biophysical evidence from size-exclusion chromatography and DLS (dynamic light scattering) indicate that trichosurin is a dimer in solution (protein concentration=0.5 mg/ml; molecular mass from DLS ∼38.6 kDa; molecular mass from size-exclusion chromatography ∼38 kDa). The surface area buried per monomer, by dimerization, is 905 Å2 (11.8% of the monomer surface, calculated from the pH 4.6 structure). At the core of the dimerization interface is the ring-to-ring packing of Tyr100 with Tyr115. These aromatic interactions are complemented further by hydrophobic burial of the side-chains of Val121 and Val123. Two salt bridge interactions between Arg150 and Asp98 also contribute to the stability of the dimer along with numerous inter-molecular hydrogen bonds. At least fifteen water molecules are present within the dimer interface in the apo-trich4.6 structure. The lower resolution of the apo-trich8.2 structure precluded the modelling of as many water molecules and, as a consequence, only four bridging water molecules are seen in the dimer interface for the higher pH structure.
A review of lipocalin dimerization modes reveals six variations in orientation. The most closely related dimer is that of porcine BLG [22], which uses part of the same face to dimerize as trichosurin (Figure 3). However, the porcine BLG dimer also has an N-terminal crossover between the monomers in which hydrogen bonding and other interactions occur, taking the effective buried surface area to a reported 1757.7 Å2 per monomer [22]. The dimerization of trichosurin is sufficiently different to that of porcine BLG that it represents a seventh variation on the theme (Figure 3).
Figure 3. The trichosurin dimer and its relationship with other lipocalin dimers.
(A) Trichosurin dimer (blue and yellow) looking down the 2-fold dimerization axis. Buried surface area upon dimerization is 905 Å2. For reference, in all cases a monomer of trichosurin is depicted in blue and yellow in the same orientation to that in (A). (B) Major horse allergen dimer (red with trichosurin monomer for reference) (PDB code 1EW3); buried surface area is 1023.3 Å2. (C) Porcine BLG dimer (yellow) (PDB code 1EXS); buried surface area is 1757.7 Å2. (D) Bovine odourant-binding protein dimer (magenta) (PDB code 1OBP); buried surface area is 2399.7 Å2. (E) Bovine BLG dimer (green) (PDB code 1B0O); buried surface area is 484.1 Å2. (F) Porcine OBP dimer (slate) (PDB code 1E06); buried surface area is 848.2 Å2; and (G) Nitrophorin 4 dimer (gold) (PDB code 1EQD); buried surface area is 789.1 Å2.
Structural comparison with lipocalins
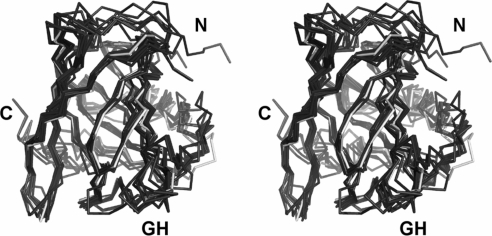
Bovine BLG is regarded as the prototypic milk lipocalin. A structural alignment of apo-trich4.8 with the BLG structure 1BEB, solved at pH 6.5, gives an rmsd of 2.76 Å over 139 equivalent residues (Figure 4, and Supplementary Table S1 and Figure S1 at http://www.BiochemJ.org/bj/408/bj4080029add.htm) and a sequence identity of 21% after structure-based sequence alignment. In general the two structures align very well, although differences are seen in the N-terminus (due to this region being undefined in the trichosurin structure) and for the GH loop (in BLG a short kinked helical structure is formed, whereas in trichosurin the same loop forms a textbook β-turn). Interestingly the EF loop is also similar between the two proteins. This loop is responsible for the pH-dependent opening and closing of the barrel in BLG; a conformational change known as the Tanford transition [23]. The determination of the structure of trichosurin at two different pHs affords an opportunity to assess the presence of an equivalent conformational change over the pH range studied. Trichosurin lacks a similar ionizable group when compared with BLG (Glu89) and does not appear to exhibit a similar opening and closing action. The orientation of the Tyr94 side chain does however change position in the higher pH structure, leading to a tighter closing of the barrel indicated by generally lower B-factors in this area of the pH 8.2 structure when compared with the main barrel. A mechanism responsible for access to the barrel interior is not immediately apparent from the current structures.
Figure 4. Comparison between trichosurin and closely related lipocalins.
A structural alignment, in stereo, of the Cα positions for trichosurin at pH 4.6 (white), with porcine odourant-binding protein (PDB code 1DZK), bovine d2 allergen (PDB code 1BJ7), hamster aphrodisin (PDB code 1E5P), equine c2 allergen (PDB code 1EW3), boar salivary lipocalin (PDB code 1GM6), mouse MUP (PDB code 1MUP), rat MUP (PDB code 2A2U), bovine BLG (PDB code 1BEB) and pig BLG (PDB code 1EXS). The N- and C-termini are labelled and GH shows the loop between β-strands G and H.
A structure–comparison survey of the SCOP [24] database places trichosurin in the ‘retinol-binding-like’ protein family. This family encompasses a total of 141 structures which include retinol-binding proteins, odourant-binding proteins, BLGs, MUPs from mice and rats, human NGAL (neutrophil gelatinase-associated lipocalin) and C8γ proteins, nitrophorins and α-crustacyanin. A multiple structure alignment carried out using MAMMOTH [25] showed that trichosurin clusters with a subset of the retinol-binding-like proteins which include porcine odourant-binding protein, the bovine d2 and equine c2 allergens, BLGs and the MUPs from mice and rats (Figure 4, and Supplementary Table S1 and Figure S1 at http://www.BiochemJ.org/bj/408/bj4080029add.htm). The majority of the structural features are well conserved among this subset with rmsd values of 1.3–3.4 Å. In contrast, the sequence identity is relatively low (16%–34%; see Supplementary Figure S2 at http://www.BiochemJ.org/bj/408/bj4080029add.htm). This points towards a general function for trichosurin as a protein which binds small ligands in the central cavity but, in the absence of tight sequence conservation, there are few clues about the precise nature of these ligands.
Ligand-binding cavity
From examination of the trichosurin structure, and from knowledge of the binding properties of other lipocalins, the central pocket inside the trichosurin barrel is expected to be the major ligand-binding site. The trichosurin-binding cavity is a hydrophobic pocket with approximate dimensions of 11×6×6 Å. The internal molecular surface, described by a probe sphere of radius 1.4 Å and using a triangulation algorithm that places six grid points every 1.4 Å [26], is 425 Å2 and encloses a total volume of ∼464 Å3. It has a bulbous top and bottom but is constricted in the centre by the side chains of Phe65 and Phe114 which protrude into the pocket (Figure 2A).
There are three asparagine residues, Asn49, Asn97 and Asn127, which lie in the cavity, and offer hydrogen-bonding sites for potential ligands. Two water molecules are bound inside the apo-trich4.6 barrel. Both are near the top of the hydrophobic pocket, with one (labelled W3 in Figure 2B) hydrogen-bonded to the carbonyl oxygen of Phe47 and the Asn127 OD1 oxygen, and the other (labelled W2 in Figure 2B) held by hydrogen bonds to Asn49 O, Asn127 OD1 and Thr30 OG1. Additionally, two regions of electron density in the cavity of apo-trich4.6 have been modelled as 2-propanol which is a constituent of the crystallization conditions. One is held by two hydrogen bonds to Asn49 OD1 and the water molecule W3 (see Figure 2C), whereas the other is buried further down in the cavity, hydrogen-bonded with Leu78 O. These 2-propanol molecules are not present in the apo-trich8.2 structure as they originate from the crystallization mother liquor.
In the apo-trich8.2 structure three water molecules are bound near the top of the pocket. Two of these water molecules occupy equivalent positions to W2 and W3 in the apo-trich4.6 structure (Figure 2B), whereas the third occupies a position in place of one of the 2-propanol molecules, bonded to Asn49 OD1 and W3. The position occupied by the 2-propanol molecule hydrogen-bonded to Leu78 O in the apo-trich4.6 structure is left unoccupied in the apo-trich8.2 structure. Water molecules found within the predominantly non-polar pocket are not unique to trichosurin. MUP has been shown to utilize internalized water molecules in the formation of water mediated ligand-to-protein hydrogen bonds [14].
There are subtle differences between the binding pockets in the apo-trich4.6 and apo-trich8.2 structures. The side chains of the 22 residues that line the binding pocket align closely. The volumes of the binding pocket at pH 4.6 and at pH 8.2 vary only slightly. This is in direct contrast with bovine BLG where significant conformational changes accompany pH changes across quite a narrow range (pH 6.2–8.2) [27].
The N-terminus
The N-terminal region, residues 1–23, is not observed in the structure determined at pH 4.6, whereas residues 1–20 are absent in the pH 8.6 structure. To ensure that the N-terminus was intact, N-terminal sequencing was performed prior to crystallization. In other lipocalins this region of the amino acid chain helps to close the bottom end of the barrel. In trichosurin, the bottom of the barrel is effectively closed by Phe89, Leu24, Phe112 and Ile54. However, in the apo-trich8.2 structure Trp21, Glu22, Gln23 and Leu24 form a classic lipocalin-like 310 helix which further protects the hydrophobic residues from the aqueous environment and no doubt plays a stabilizing role in this region of the structure.
In structures of MUP, part of the N-terminus is often disordered, but this only accounts for the first five residues, with the rest of the N-terminal region hydrogen bonding to the kinked end of β-strand F in pseudo β-sheet fashion before turning to pass under the bottom of the barrel and form the 310 helix. The trichosurin N-terminal residues do not follow the same path. At this stage it is not possible to assign structure or a role to these 20 residues; however, they may perform a specific function such as a receptor-binding interaction whereupon they might assume a definite structure upon complexation with a suitable receptor. Such an interaction is known for the C-terminus of plasma retinol-binding protein, a lipocalin, upon formation of its complex with transthyretin [28].
Bound ligands
There have been many studies on ligand binding by various lipocalin family members, such as BLG [29], retinol-binding protein [30], MUP [14,31] and various human lipocalins [32]. In some cases, for example MUP, these studies have proven useful in the determination of probable function, but, in others, such as BLG, the information has failed to offer clear insights into biological function.
Given the close structural similarity of trichosurin to MUP, and initial hypotheses surrounding pouch young retention and possible pheromone communication [10], six ligands known to bind to MUP were selected to investigate pocket binding specificity in trichosurin (Figure 5). The six potential ligands thymol, trans-2-nonenal, 2-naphthol, 4-ethylphenol, 2-sec-butyl-thiazole and 2-isobutyl-3-methoxypyrazine were used as molecular probes. Ligand screening of six potential small molecules was carried out by co-crystallization of trichosurin in the presence of 3–5 mM ligand. With the exception of 2-sec-butyl-thiazole, structures were solved for each of the co-crystallizations and this showed that, of the five ligands tested, two bound in the pocket of trichosurin: 2-naphthol and 4-ethylphenol. For the remaining three ligands, no electron density could be seen in the trichosurin-binding pocket.
Figure 5. Potential ligands for trichosurin.
The chemical structures for six ligands which were assessed for binding to trichosurin in vitro using fluorescence and crystallography (a–f) and two ligands assessed using in silico docking (g and h).
The 2-naphthol ligand binds in the top and centre of the pocket and makes a hydrogen bond to an internal water molecule, which in turn, is hydrogen-bonded by Phe47 O, Asn127 OD1 and Asn49 ND1 (see Figure 2B). Despite the presence of the ligand, the barrel remains completely closed. The binding of this ligand has resulted in a minor side chain rearrangement inside the binding pocket. The plane of the aromatic ring of Phe114 is rotated such that a small cavity is formed between the ring and the amide side chain of Asn127 and this is occupied by a water molecule. The water molecule (W232 in Figure 2B) accepts a hydrogen bond from the amide nitrogen but is also close enough to the aromatic ring to form an unconventional hydrogen bond by donation to the delocalized electron density of the aromatic ring plane. This phenomenon has been previously observed and described [33].
In contrast with the binding of 2-naphthol, 4-ethylphenol binds in the lower of the two lobes of the binding pocket forming a hydrogen bond to Leu78 O. The axis of the ligand is oriented perpendicular to the barrel axis (see Figure 2C), which is also in contrast with the binding of 2-naphthol. With the ligand in this position the trichosurin-binding pocket is only partially filled and in this structure a 2-propanol solvent molecule is able to occupy the upper lobe of the binding pocket. The trichosurin barrel is again completely closed.
In addition to the ligand electron density within the trichosurin barrel, the 2-naphthol structure also shows some electron density in a region bounded by the external barrel wall, the N-terminal helix and the dimer interface (see Figure 2D). This region of electron density lies in a trough formed by these three elements and is enclosed on a fourth side by the side chain of Arg150. It was designated as a second, externally bound 2-naphthol molecule, making mainly hydrophobic interactions and also a favourable aromatic interaction with the delocalized electrons of the guanidine head group of Arg150. In this position the 2-napthol molecule remains partially solvent exposed with 6–12 Å2 accessible to the bulk solvent. The occupancy for this second binding site has been refined at 0.7 and is lower than that found for the main binding site of 0.9.
X-ray crystallographic studies on crystals grown in the presence of three of the four remaining possible ligands (trans-2-nonenal, thymol and 2-isobutyl-3-methoxy pyrazine) indicated that these molecules did not bind to trichosurin in the main pocket or externally. No suitable crystals were grown in the presence of 2-sec-butyl thiazole.
Fluorescence titration data were collected for five potential ligands binding to trichosurin: 2-napthol, 4-ethylphenol, trans-2-nonenal, thymol and 2-isobutyl-3-methoxy pyrazine (Figures 5a–5e). Curves were fitted to the data for 2-naphthol following the change in fluorescence at 326 nm (Figure 6) and binding constants were calculated using a simple model for consecutive binding; P+L↔PL+L↔PL2 (where P is protein and L is ligand). This ignores complicating factors such as cooperativity between binding sites both within individual binding sites and between monomers of the dimer, as well as the dimerization Kd. The data are also convoluted by the overlapping fluorescence of both the ligand and the protein, and the dramatic change in pKa for the ligand before and after excitation [34] especially with respect to the differing local pH effects caused by binding in the two sites. This precludes using more complicated models. For these reasons, data were recorded at emission wavelengths below the emission spectrum of the ligand (<330 nm).
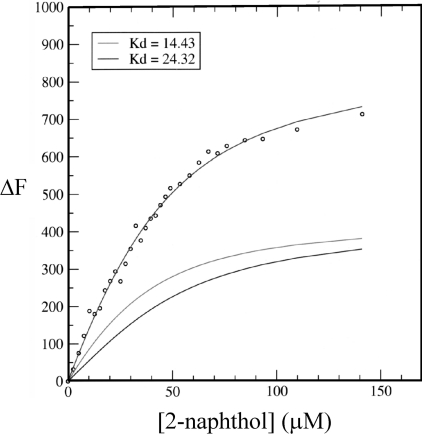
Figure 6. Curves fitted for ΔF at 326 nm with respect to ligand concentration.
The change in fluorescence at 326 nm (ΔF) for ligand binding to trichosurin. ΔF on the y-axis is in arbitrary units. The upper line is the best fit of two exponentials to the data (○) with Kd values of 14.4 (middle line) and 24.3 μM (lower line).
For the change in fluorescence at 326 nm the curve fitting resulted in a Kd of 14 μM for the stronger site and of 24 μM for the weaker site. These binding data make sense in the context of the structure where binding into the cavity is expected to be favoured over binding in a secondary solvent-exposed site. Changes in fluorescence for 4-ethylphenol binding to trichosurin were followed at 320 nm where the intrinsic fluorescence of the ligand is at a minimum and the fluorescence of the protein is near its maximum. The amplitude of the changes in fluorescence were moderately small and are practically linear for ligand conentrations up to 60 μM. This makes the data difficult to interpret; however, it sets a lower bound for the Kd for 4-ethylphenol of 30 μM with the possibility that this Kd is much weaker.
Fluorescence spectra for titrations with trans-2-nonenal, thymol and 2-isobutyl-3-methoxy pyrazine showed no changes in protein fluorescence for these compounds. This indicates that either these compounds are binding to trichosurin and not affecting the fluorescence of the protein or that they are not binding. The latter case is supported by the crystallographic data which shows no binding to trichosurin even at concentrations of 3–5 mM ligand. These data indicate that trichosurin preferentially binds small phenolic compounds. Furthermore, the preference for 2-napthol and 4-ethylphenol over thymol (Figures 5a, 5b and 5f respectively) indicates a specificity for phenolic compounds with particular substituents around the aromatic ring.
In comparison with other lipocalins, the Kd for the binding of 2-naphthol to MUP is 0.38 μM [35] and Kd values for a group of four ligands [retinol, retinoic acid, DAUDA {11-[5-(dimethylamino)-1-naphthalenesulfonylamino]undecanoic acid} and ANS (8-anilino-1-naphthalenesulfonic acid)] binding to ten different human lipocalins range from 0.1–10 μM [32]. The Xenopus laevis coroid plexus lipocalin Cp11 binds retinal in the nanomolar range and retinoic acid in the micromolar range [36,37].
The specificity of trichosurin for 2-naphthol and 4-ethylphenol with relatively high Kd values suggests that small phenolic compounds of this type may have higher affinities and constitute the natural ligand for trichosurin. This is the subject of an ongoing screen and some possible suggestions are made in the subsequent discussion.
In summary, the structure of trichosurin is that of an eight-stranded β-barrel typical of the lipocalins. The binding of two solvent-derived isopropanol molecules within the putative trichosurin-binding pocket is suggestive of the manner in which the natural substrate or substrates may be bound. The shape and charge distribution within the pocket indicate that a small hydrophobic molecule perhaps with substituent groups may easily be accommodated within the closed pocket, with sites provided by the protein to satisfy potential hydrogen bonding requirements. This is confirmed by the specific binding of 4-ethylphenol and 2-naphthol over other possible ligands. It is unclear at this point what, if any, mechanism is required to enable access to the binding pocket for molecules larger than 2-propanol. They may well enter and exit through the N-terminal end of the barrel given the disorder seen in the trichosurin structure in this region, although this would be contrary to established theories about lipocalin ligand binding. There are few differences between the structures of trichosurin solved from crystals grown at two different pH values. The overall mode of dimerization is consistent between the two structures and does not appear to show a dependence on pH as is the case for bovine BLG. Furthermore, at the two pH values investigated, trichosurin did not show a major conformational difference consistent with the pH-induced barrel opening and closing seen for BLG [38]. It is possible that such an opening and closing mechanism could occur outside of the pH range studied. The biological purpose, if any, of a bound Zn2+ ion on the barrel exterior remains unknown, although it could be involved in orientation and binding during a receptor interaction.
Trichosurin is a highly conserved protein found in the milk of metatherians but its role in the milk is not understood. It is most closely related to the MUPs (from mice and rats) whose name is somewhat historical as the primary site of expression of these proteins is the liver and they have also been shown to be expressed in many other tissues such as the submaxiliary, lachrymal, parotid and mammary glands where they play a very diverse set of functions. From the similarity between trichosurin and the MUPs, it is tempting to speculate that the role of trichosurin might be in delivering attractants to guide the precarious path taken by the foetus from uterus to nipple in the early stages of marsupial development. However, trichosurin is a significant constituent of milk throughout possum lactation [39] and this argues against a chemical attractant hypothesis.
A second hypothesis regarding the putative function of trichosurin is suggested by the demonstrated preference that we see for small phenolic ligands. Phenolic compounds are a significant constituent in plants and they are utilized by the plants as toxic feeding deterrents. These compounds have a very significant effect on feeding behaviour among mammalian herbivores as the animals ability to metabolize (detoxify) these compounds directly limits dietary intake. This has been demonstrated to be the case in possums [40]. Furthermore, female possums are able to concentrate phenolic compounds in their milk to levels appreciably higher than those found in the blood plasma [41]. This provides the young with a source of phenolics to enable priming of the neonate liver in the production of detoxifying enzymes which possums are known to utilize to allow them to survive on otherwise toxic foliage. The specificity for small phenolics that we observe in the present study is consistent with the hypothesis that trichosurin may facilitate the process of carrying secondary plant phenolics through the milk and help in transfer across the neonate gut. We have used the program GOLD [42] to dock common plant phenolics such as cinneol and gallic acid (Figures 5g and 5h) into the ligand-binding pocket of trichosurin and this shows that these ligands are of a size and have hydrogen-bonding groups which make them compatible ligands for trichosurin. As all the marsupials are generalist herbivores, this hypothesis of phenolic priming of detoxification pathways is consistent with the high level of sequence conservation of trichosurin among the metatherians (from the South American opossum to the tammar wallaby). We have also used GOLD to survey other possible ligands and we note that planar-fused benzene rings analogous to 2-naphthol, such as 2-phenathrenol and 2-hydroxyanthracene, can also be accommodated sterically and chemically (with good GOLD scores) by the ligand-binding pocket. This may point to a more promiscuous binding function for trichosurin.
We have determined the structures of the first metatherian lipocalin which is present at high levels throughout lactation in the possum, pointing towards an important role in the unusual development of the marsupial foetus. Marsupials occupy a unique position on the evolutionary tree as they diverged from the mammals midway between the divergence of placental mammals and the divergence of mammals from other animals such as birds. We have shown a preference for small phenolic ligands by trichosurin which bind with micromolar affinity and provides a possible functional hypothesis. However, the crucial next step is to discover the natural ligands for trichosurin and this is the subject of ongoing work.
Online data
Acknowledgments
We would like to thank M. Grigor (Faculty of Science, University of Auckland, Auckland, New Zealand) for the donation of the original trichosurin cDNA clone and for discussions and critical reading of the manuscript, and Dr Didier Nurizzo for help with synchrotron data collection at the ESRF, and Dr Armin Widmer for custom curve-fitting algorithms. We acknowledge financial support from the Marsden Fund of New Zealand.
References
- 1.Cowan P. E. Changes in milk composition during lactation in the common brushtail possum, Trichosurus vulpecula (Marsupialia: Phalangeradae) Reproduct. Fertil. Dev. 1989;1:322–333. doi: 10.1071/rd9890325. [DOI] [PubMed] [Google Scholar]
- 2.Basse B., Flux I., Innes J. Recovery and maintenance of North Island Kokako (Callaeas cinerea wilsoni) populations through pulsed pest control. Biol. Conserv. 2003;109:259–270. [Google Scholar]
- 3.Payton I. J., Forester L., Frampton C. M., Thomas M. D. Response of selected tree species to culling of introduced Australian brushtail possums Trichosurus vulpecula at Waipoua Forest, Northland, New Zealand. Biol. Conserv. 1997;81:247–255. [Google Scholar]
- 4.Nugent G., Fraser W., Sweetapple P. Top down or bottom up?. Comparing the impacts of introduced arboreal possums and ‘terrestrial’ ruminants on native forests in New Zealand. Biol. Conserv. 2001;99:65–79. [Google Scholar]
- 5.Sharman G. B. The initiation and maintenance of lactation in the marsupial, Trichosurus vulpecula. J. Endocrinol. 1962;25:375–385. doi: 10.1677/joe.0.0250375. [DOI] [PubMed] [Google Scholar]
- 6.Grigor M. R., Bennet B. L., Carne A., Cowan P. E. Whey proteins of the common brushtail possum (Trichosurus vulpecula): isolation, characterisation and changes in concentration during lactation of transferrin, α-lactalbuimn and serum albumin. Comp. Biochem. Physiol. 1991;98:451–459. doi: 10.1016/0305-0491(91)90239-a. [DOI] [PubMed] [Google Scholar]
- 7.Demmer J., Ross I. K., Ginger M. R., Piotte C. K., Grigor M. R. Differential expression of milk protein genes during lactation in the common brushtail possum (Trichosurus vulpecula) J. Mol. Endocrinol. 1998;20:37–44. doi: 10.1677/jme.0.0200037. [DOI] [PubMed] [Google Scholar]
- 8.Demmer J. The prolactin receptor from the brushtail possum (Trichosurus vulpecula): cDNA cloning, expression and functional analysis. Mol. Cell. Endocrinol. 1999;148:119–127. doi: 10.1016/s0303-7207(98)00230-5. [DOI] [PubMed] [Google Scholar]
- 9.Piotte C. P., Marshall C. J., Hubbard M. J., Collet C., Grigor M. R. Lysozyme and α-lactalbumin from the milk of a marsupial, the common brush-tailed possum (Trichosurus vulpecula) Biochim. Biophys. Acta. 1997;1336:235–242. doi: 10.1016/s0304-4165(97)00033-0. [DOI] [PubMed] [Google Scholar]
- 10.Piotte C. P., Hunter A. K., Marshall C. J., Grigor M. R. Phylogenetic analysis of three lipocalin-like proteins present in the milk of Trichosurus vulpecula (Phalangeridae, Marsupiala) J. Mol. Evol. 1998;46:361–369. doi: 10.1007/pl00006313. [DOI] [PubMed] [Google Scholar]
- 11.Flower D. R. The lipocalin protein family: structure and function. Biochem. J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akerstrom B., Flower D. R., Salier J.-P. Lipocalins: unity in diversity. Biochim. Biophys. Acta. 2000;1482:1–8. doi: 10.1016/s0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- 13.Sawyer L., Kontopidis G. The core lipocalin, bovine β-lactoglobulin. Biochim. Biophys. Acta. 2000;1482:136–148. doi: 10.1016/s0167-4838(00)00160-6. [DOI] [PubMed] [Google Scholar]
- 14.Timm D. E., Baker L. J., Mueller H., Zidek L., Novotny M. V. Structural basis of pheromone binding to mouse major urinary protein (MUP-I) Protein Sci. 2001;10:997–1004. doi: 10.1110/ps.52201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flower D. R., North A. C. T., Sansom C. E. The lipocalin protein family: structural and sequence overview. Biochim. Biophys. Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 16.Jancarik J., Kim S. H. Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr. 1991;24:409–411. [Google Scholar]
- 17.Otwinowski Z., Minor W. Denzo and Scalepack. Methods Enzymol. 1997;267:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 19.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple refined dummy atomic models. Acta Crystallogr. 1997;D53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 20.Jones A. T. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse K. R., Jiang J. S., Kuszewski J., Simonson T., Warren G. L. Crystallography and NMR System, CNS, a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Hoedemaeker F. J., Visschers R. W., Alting A. C., de Kruif K. G., Kuil M. E., Abrahams J. P. A novel pH-dependent dimerisation motif in β-lactoglobulin from pig (Sus scrofa) Acta Crystallogr. 2002;D58:480–486. doi: 10.1107/s0907444902000616. [DOI] [PubMed] [Google Scholar]
- 23.Tanford C., Lyle G. B., Yashohiko N. The reversible transformation of β-lactoglobulin at pH 7.5. J. Am. Chem. Soc. 1959;81:4032–4035. [Google Scholar]
- 24.Andreeva A., Howorth D., Brenner S. E., Hubbard T. J. P., Chothia C., Murzin A. G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupyan D., Leo-Macias A., Ortiz A. R. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics. 2005;21:3255–3263. doi: 10.1093/bioinformatics/bti527. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt G. J., Jones T. A. Databases in protein crystallography. Acta Crystallogr. 1998;D54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 27.Qin B. Y., Bewley M. C., Creamer L. K., Baker H. M., Baker E. N., Jameson G. B. Structural basis of the tanford transition of bovine β-lactoglobulin. Biochemistry. 1998;37:14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 28.Newcomer M. E., Ong D. E. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim. Biophys. Acta. 2000;1482:57–64. doi: 10.1016/s0167-4838(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 29.Sawyer L., Brownlow S., Polikarpov I., Wu S.-Y. β-Lactoglobulin: structural studies, biological clues. Int. Dairy J. 1998;8:65–72. [Google Scholar]
- 30.Monaco H. L., Zanotti G. Three-dimensional structure and active site of three hydrophobic molecule-binding proteins with significant amino acid sequence similarity. Biopolymers. 1992;32:457–465. doi: 10.1002/bip.360320425. [DOI] [PubMed] [Google Scholar]
- 31.Lehman-McKeeman L. D., Caudill D., Rodriguez P. A., Eddy C. 2-sec-butyl-4,5-dihydrothiazole is a ligand for mouse urinary protein and rat α 2u-globulin: physiological and toxicological relevance. Toxicol. Appl. Pharmacol. 1998;149:32–40. doi: 10.1006/taap.1997.8343. [DOI] [PubMed] [Google Scholar]
- 32.Breustedt D. A., Schonfeld D. L., Skerra A. Comparative ligand-binding analysis of ten human lipocalins. Biochim. Biophys. Acta. 2006;1764:161–173. doi: 10.1016/j.bbapap.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Steiner T. Hydrogen bonds from water molecules to aromatic acceptors in very high-resolution protein crystal structures. Biophys. Chem. 2002;95:195–201. doi: 10.1016/s0301-4622(01)00256-3. [DOI] [PubMed] [Google Scholar]
- 34.Trieff N. M., Sundheim B. R. The effect of solvent and acid-base kinetics of the excited state of 2-naphthol. J. Phys. Chem. 1965;69:2044–2059. [Google Scholar]
- 35.Sartor G., Pagani R., Ferrari E., Sorbi R. T., Cavaggioni A., Cavatorta P., Spisni A. Determining the binding capability of the mouse major urinary proteins using 2-naphthol as a fluorescent probe. Anal. Biochem. 2001;292:69–75. doi: 10.1006/abio.2001.5065. [DOI] [PubMed] [Google Scholar]
- 36.Lepperdinger G. Amphibian choroid plexus lipocalin, Cp11. Biochim. Biophys. Acta. 2000;1482:119–126. doi: 10.1016/s0167-4838(00)00143-6. [DOI] [PubMed] [Google Scholar]
- 37.Lepperdinger G., Strobl B., Jilek A., Weber A., Thalhamer J., Flockner H., Mollay C. The lipocalin Xlcpl1 expressed in the neural plate of Xenopus laevis embryos is a secreted retinaldehyde binding protein. Protein Sci. 1996;5:1250–1260. doi: 10.1002/pro.5560050704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jameson G. B., Adams J. J., Creamer L. K. Flexibility, functionality and hydrophobicity of bovine β-lactoglobulin. Int. Dairy J. 2002;12:319–329. [Google Scholar]
- 39.Demmer J., Stasiuk S. J., Grigor M. R., Simpson K. J., Nicholas K. R. Differential expression of the whey acidic protein gene during lactation in the brushtail possum (Trichosurus vulpecula) Biochim. Biophys. Acta. 2001;1522:187–194. doi: 10.1016/s0167-4781(01)00334-7. [DOI] [PubMed] [Google Scholar]
- 40.Dearing M. D., Cork S. Role of detoxification of plant secondary compounds on diet breadth in a mammalian herbivore, Trichosurus vulpecula. J. Chem. Ecol. 1999;25:1205–1219. [Google Scholar]
- 41.MacLennan D. G., Blume L. M., Johnson D. E. Induction of phenol detoxifying enzymes in Juvenile brushtail possums Trichosurus vulpecula (marsupialia) by phenols transmitted in the maternal milk. Toxicon. 1983;S3:261–264. [Google Scholar]
- 42.Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. Improved protein–ligand docking using GOLD. Proteins Struct. Funct. Genet. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.