Abstract
Sirt1 is an NAD+-dependent deacetylase that plays a role in cellular processes such as transcriptional regulation, stress response, longevity and apoptosis. Sirt1 deacetylates histone proteins and certain transcription factors such as p53, CTIP2 (chicken ovalbumin upstream promoter-transcription factor-interacting protein 2), FOXO (forkhead box O) and NF-κB (nuclear factor κB). To identify potential Sirt1-interacting factors, we performed a yeast two-hybrid screen. The screen identified TLE1 (transducin-like enhancer of split-1) as a possible Sirt1-interacting factor, which was then confirmed by co-immunoprecipitation. TLE1 is a non-DNA binding co-repressor for several transcriptional factors including NF-κB. We have demonstrated using co-transfection assays that Sirt1 and TLE1 repress NF-κB activity. The catalytic mutant of Sirt1, Sirt1-H363Y, and the N-terminal Sirt1 fragment (amino acids 1–270) also show similar repression activity, suggesting that the deacetylase activity of Sirt1 may not be critical for its effect on NF-κB activity. Furthermore, analysis in Sirt1-null MEFs (murine embryonic fibroblasts) and HeLa cells stably expressing siRNA (small interfering RNA) specific to Sirt1 or TLE1 demonstrate that both Sirt1 and TLE1 are required for negative regulation of NF-κB activity. Taken together, these results suggest that the interaction between Sirt1 and TLE1 is important for mediating repression of NF-κB activity.
Keywords: deacetylase, nuclear factor κB (NF-κB), Sirt1, transducin-like enhancer of split-1 (TLE1), transcription
Abbreviations: AD, activation domain; AES, N-terminal enhancer of split; BCA, breast-cancer-associated protein; bHLH, basic helix–loop–helix; CTIP, chicken ovalbumin upstream promoter-transcription factor-interacting protein; DBD, DNA-binding domain; FOXO, forkhead box O; HDAC, histone deacetylase; IκB, inhibitory κB; IL-8, interleukin 8; LB, Luria–Bertani; MEF, murine embryonic fibroblast; NF-κB, nuclear factor κB; PCAF, p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein]-associated factor; PPAR-γ, peroxisome-proliferator-activated receptor-γ; PGC1α, PPARγ co-activator 1α; SD medium, selective drop-out medium; siRNA, small interfering RNA; TLE1, transducin-like enhancer of split-1; TNFα, tumour necrosis factor α; X-gal, 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside
INTRODUCTION
Sirt1, an NAD+-dependent deacetylase, is the human homologue of the yeast Sir2 protein implicated in transcriptional regulation, DNA damage response, cell growth arrest, cellular metabolism and apoptosis. Sirt1 associates with the bHLH (basic helix–loop–helix) repressor proteins HES1 (hairy and enhancer of split-1), HEY2 (hairy and enhancer of split-related with YRPW motif 2) [1] and CTIP2 (chicken ovalbumin upstream promoter-transcription factor-interacting protein 2) [2] to confer transcriptional repression. In addition, Sirt1 deacetylates non-histone substrates such as p53, PCAF {p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein]-associated factor}, MyoD, p300, Ku70, PPARγ (peroxisome-proliferator-activated receptor γ) and PGC1α (PPARγ co-activator 1α) [3–8]. Sirt1 also increases cellular resistance to DNA damage by inhibiting p53-mediated apoptosis [9]. However, Sirt1 can also deacetylate p53 without affecting cell survival following DNA damage [10], suggesting the possibility of deacetylase-independent functions of Sirt1 under certain cellular stress conditions. Sirt1 also increases cellular resistance to oxidative stress by a p53-independent pathway [11,12], suggesting that there are multiple pathways through which Sirt1 regulates the cellular response to apoptotic challenge.
NF-κB (nuclear factor κB) is a transcription factor that regulates the expression of a variety of genes involved in critical cellular processes ranging from the immune response to carcinogenesis. The vertebrate Rel/NF-κB family comprises five proteins, c-Rel, RelA, RelB, p105/NF-κB and p100/NF-κB, that are structurally related. Transcriptionally active NF-κB consists of a heterodimeric protein complex with, most commonly, the p50–RelA/p65 heterodimer. Acetylation of p65 at Lys122 and Lys123 decreases DNA-binding affinity of the heterodimer, whereas acetylation at Lys310 and Lys221 increases DNA binding and impairs its association with IκB (inhibitory κB) [13]. Members of the Class I histone deacetylases, HDAC1, HDAC2 and HDAC3, are known to regulate the activity of NF-κB by deacetylating RelA/p65, increasing its association with IκB and reducing transcriptional activity. Sirt1 has also been reported to repress NF-κB-mediated gene expression by deacetylating the p65 subunit of NF-κB [14].
NF-κB activity is also regulated by members of the TLE1 (transducin-like enhancer of split-1) family of transcriptional repressors, which are mammalian homologues of the Drosophila Groucho protein family. Groucho binds directly to the Drosophila homologue of NF-κB, Dorsal, and converts it from a transcriptional activator into a repressor [15]. The mammalian Groucho homologues TLE1 and the AES (N-terminal enhancer of split) also physically interact with the p65 subunit of NF-κB to inhibit its activity through an unknown mechanism [16].
To identify potential Sirt1-interacting protein partners, we performed a yeast two-hybrid screen using a GAL4–Sirt1 fusion as bait. We identified TLE1 as a potential factor that was able to interact with Sirt1, an association that was confirmed in mammalian cells by co-immunoprecipitation. In addition, we demonstrated that repression of NF-κB activity mediated by Sirt1 and TLE1 was enhanced when both of the proteins are overexpressed. More significantly, neither Sirt1 nor TLE1 was able to repress NF-κB activity in the absence of the other. Interestingly, our results suggest that the catalytic function of Sirt1 was not required for the repression of NF-κB. These results suggest that the association between Sirt1 and TLE1 plays a critical role in mediating Sirt1- and TLE1-regulated repression of NF-κB activity and possibly the activity of other transcription factors.
MATERIALS AND METHODS
Construction of plasmids for yeast two-hybrid screen
A point mutation was carried out using the QuikChange® site-directed mutagenesis kit (Stratagene) to create an in-frame BamHI/SalI cloning site in the yeast expression vector pGBKT7 (Clontech Laboratories) using the primers 5′-GGAGGCCGAATTCCCGGGATCCGTCGACCTGCAG-3′ and 5′-CTGCAGGTCGACGGATCCCGGGAATTCGGCCTCC-3′. A total of 12 cycles of PCR using PfuTurbo DNA polymerase (Stratagene) was performed as per the manufacturer's protocol. The PCR product was digested with the restriction enzyme DpnI to eliminate the parental supercoiled methylated dsDNA. The PCR-synthesized plasmids were then transformed into XL-Blue supercompetent Escherichia coli cells, and plasmids were purified using a Maxiprep Kit (Qiagen). The full-length Sirt1 cDNA was obtained from pYESir2-puro, a gift from Robert Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA, U.S.A.) [9], and ligated in-frame into the BamHI/SalI GAL4 DBD (DNA-binding domain) of the frameshifted yeast expression vector to generate GAL4-DBD–Sirt1 (pGBKT7-Sirt1) for the yeast two-hybrid screen.
Yeast two-hybrid screen
Yeast two-hybrid screening was performed according to the Clontech Matchmaker protocol using the full-length Sirt1 as bait and the human spleen cDNA library (Clontech Laboratories) as prey. Briefly, the cDNA library was first titred and then amplified to isolate 10.5×106 (three times the size of the library) independent colonies. The library was plated on LB (Luria–Bertani) plus ampicillin plates at a high enough density so as to get approx. 40000 colonies per plate (fully confluent 150 mm plates) using 5 mm diameter sterile glass beads. Plates were incubated at 37 °C for 18–20 h. A 5 ml aliquot of sterile LB plus glycerol was added to each plate and the colonies were scraped off using a glass spreader. The pooled colonies were incubated at 30 °C for 3 h with shaking at 200 rev./min. Library plasmid was extracted using a Megaprep kit (Qiagen).
AH109 yeast cells were transformed with pGBKT7-Sirt1 using the PLATE {PEG [poly(ethylene glycol)]/lithium acetate/Tris/EDTA} transformation protocol (Clontech Matchmaker) for verification of fusion protein expression. Transformants were selected on SD (selective drop-out medium)−Trp plates. Protein extraction from yeast was performed using an SDS/urea method (Yeast Protocol Handbook, Clontech). Western blot analysis was performed on the extracted protein using a monoclonal anti-Sirt1 antibody (Upstate). The large-scale simultaneous co-transformation protocol was performed according to the manufacturer's instructions (Clontech). A total of 50 μg of the human cDNA expression library fused to the GAL4 transactivation domain in the pACT2 vector (Clontech) [AD (activation domain)/library] and 100 μg of the full-length Sirt1 fused to the GAL4 DBD in the pGBKT7 yeast expression vector (DBD/bait) was used. Co-transformants were selected on high-stringency SD−His/−Ala/−Leu/−Trp plus 2.5 mM aminotriazol plates at 30 °C. Plasmids expressing GAL4-DBD–p53 and GAL4-AD–T-antigen were used as positive controls, whereas lamin C and T-antigen were used as negative controls. The 78 colonies that grew on high-stringency selection plates were streaked on SD/−Lys/−Trp/plus X-gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside) plate for the β-galactosidase assay for further confirmation of positive clones. Yeast plasmids were rescued from clones that were positive for β-galactosidase activity using the Hoffman and Winston method (Clontech Laboratories) and were analysed by nucleotide sequencing. The cDNA sequences were compared with the NCBI (National Center for Biotechnology Information) GenBank® database for identification of the interacting proteins.
Cell culture, transfection and retroviral transduction
HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were transfected at 70–80% confluency using the Lipofectamine™ 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol. At 24 h post-transfection, the cells were treated with 10 ng/ml TNFα (tumour necrosis factor α) (R&D Systems) for 3 h. Cells were harvested followed by protein extraction using Nonidet P40 lysis buffer. Luciferase assays were performed using the Luciferase Assay System (Promega). Renilla luciferase activity expressed from the pRL-TK plasmid (Promega) was used as an internal control for transfection efficiency. The results are presented as the percentage luciferase activity (means±S.E.M.) for three independent transfections relative to the control.
siRNA (small interfering RNA) was used to deplete Sirt1 or TLE1 from HeLa cells by using retroviral infection. Synthetic oligonucleotide sequences 5′-GATCCGCGTGCCAAACAGGTGACCATTCAAGAGATGGTCACCTGTTTGGCACGCTTTTTTTCTAGAG-3′ and 5′-GATCCGCTGGAGCTGGGGTGTCTGTTTCAAGAGAACAGACACCCCAGCTCCAGCTTTTTTTCTAGAG-3′ (italics indicate restriction enzyme overhangs; bold indicates 19 nucleotide RNA target sense and antisense sequences) [26,27] were cloned into the retroviral vector pSirenRetroQ to create retroviral plasmids expressing siRNA for TLE1 (pSirenRetroQ-TLE1) and Sirt1 (pSirenRetroQ-Sirt1) respectively. HEK-293T cells [human embryonic kidney cells expressing the large T-antigen of SV40 (simian virus 40)] were transfected with pSirenRetroQ-TLE1, pSirenRetroQ-Sirt1 or pSirenRetroQ-negative control siRNA (BD Biosciences), along with the amphotropic envelope packaging plasmid (5 μg) using Lipofectamine™ 2000 transfection reagent according to the manufacturer's protocol. The medium, containing retroviruses, was collected 48 h after transfection and centrifuged at 1000 g for 5 min. The supernatant was then used to infect the target HeLa cells supplemented with polybrene (4 μg/ml) in a 37 °C incubator for 48 h. Infected cells were selected with puromycin (1 μg/ml) for 7 days. Protein expression was analysed by Western blotting using anti-TLE1 polyclonal antibody (Santa Cruz Biotechnology) and anti-Sirt1 monoclonal antibody (Upstate).
Co-immunoprecipitation assays
HeLa cells were transfected with plasmids expressing Myc-tagged TLE1 and FLAG-tagged Sirt1. Whole-cell protein was extracted with Nonidet P40 lysis buffer followed by protein quantification using the Bradford method. Whole-cell extract containing 500 μg of protein was incubated with 1 μg of anti-Sirt1 mouse monoclonal antibody (Upstate) or anti-TLE1 rabbit polyclonal antibody (Upstate) overnight at 4 °C. As a control, mouse IgG (Santa Cruz Biotechnology) was used. Then, 30 μl of a 50% Protein G–Sepharose bead slurry was added and incubated for 1.5 h at 4 °C. The beads were then washed five times with 1 ml of lysis buffer. Antibody-bound complexes were eluted by boiling the beads in 1×SDS protein loading buffer. Proteins were resolved by SDS/PAGE (12% gel) and transferred on to PVDF membrane (Millipore) which was then probed with anti-Myc antibody and anti-FLAG monoclonal antibody. The immunoreactive proteins were visualized by enhanced chemiluminescence (PerkinElmer Life Science).
For endogenous protein–protein interaction studies, whole-cell extract from HeLa cells was used for co-immunoprecipitation as described above using the anti-TLE1 polyclonal antibody or anti-Sirt1 monoclonal antibody for immunoprecipitation and the anti-Sirt1 monoclonal antibody or anti-TLE1 polyclonal antibody for Western blot analysis.
RESULTS
Sirt1 interacts with TLE1 in vivo
To identify potential binding partners of Sirt1, we used a yeast two-hybrid system with Sirt1 fused to the GAL4 DBD as the bait. We screened approx. 10.5×106 transformants of a human spleen cDNA library fused to the GAL4 AD in the AH109 yeast strain. A high-stringency nutritional selection medium (SD−His/−Ala/−Leu/−Trp plus 2.5 mM aminotriazol) was used for the primary selection of positive clones. Two of the positive clones were shown to encode TLE1. The identification of TLE1 as a binding partner of Sirt1 in the yeast two-hybrid screen was of particular interest because both Sirt1 and TLE1 are known to negatively regulate NF-κB-mediated transcription.
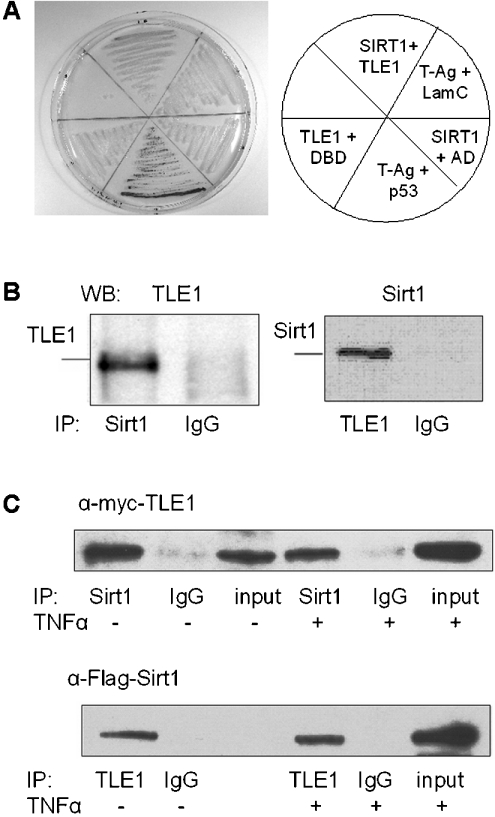
To confirm further the interaction between Sirt1 and TLE1, the plasmids expressing the GAL4-AD–TLE1 fusion, rescued from two positive yeast two-hybrid clones, were reintroduced into the Y187 yeast strain along with the plasmid expressing GAL4-DBD–Sirt1. Protein–protein interaction was demonstrated by the development of blue colouration on X-gal plates. As shown in Figure 1(A), blue colonies (image in greyscale) were observed only with colonies expressing Sirt1–DBD and TLE1–AD and the positive control consisting of plasmids expressing DBD–p53 and AD–T-antigen, but not with the controls consisting of plasmids expressing Sirt1 and AD, DBD and TLE1 or DBD–lamin C and AD–T-antigen.
Figure 1. Sirt1 and TLE1 interact in mammalian cells.
(A) Confirmation of Sirt1–TLE1 interaction in yeast two-hybrid screen. Colour change of strain Y187 on SD−Lys/−Trp plus X-gal plates after transformation with DBD–Sirt1 and AD–TLE1 plasmid rescued from a positive clone in the β-galactosidase assay. Other control sets are as follows: plasmids expressing AD–TLE1 and DBD, DBD–Sirt1 and AD, DBD–T-antigen and AD–p53, DBD–T-antigen and AD–lamin C. (B) Whole-cell extracts from HeLa cells were immunoprecipitated (IP) with anti-Sirt1 and anti-TLE1 antibodies. For control, the extracts were immunoprecipitated with mouse monoclonal IgG and rabbit polyclonal IgG respectively. The immunoprecipitated proteins were immunoblotted (WB) with anti-TLE1 and anti-Sirt1 antibodies respectively. (C) HeLa cells were transfected with Myc–TLE1 and FLAG–Sirt1 and whole-cell extracts were prepared 24 h post-transfection with or without TNFα induction. Sirt1 and TLE1 were immunoprecipitated (IP) using anti-Sirt1 and anti-TLE1 antibody respectively, followed by Western blotting using anti-Myc and anti-FLAG antibodies respectively.
To demonstrate that Sirt1 interacts with TLE1 in mammalian cells, the ability to co-immunoprecipitate endogenous Sirt1 and TLE1 as well as transiently expressed epitope-tagged versions of the proteins was examined. Immunoprecipitation of whole-cell extracts from HeLa cells with anti-TLE1 or anti-Sirt1 antibodies followed by Western blot analysis with anti-Sirt1 or anti-TLE1 antibody respectively, demonstrated that the endogenous Sirt1 and TLE1 proteins interact under physiological conditions (Figure 1B). Consistent with this observation, FLAG-tagged Sirt1 and Myc-tagged TLE1 were also co-immunoprecipitated from transfected HeLa cells (Figure 1C).
Sirt1 and TLE1 repress NF-κB-mediated transcription
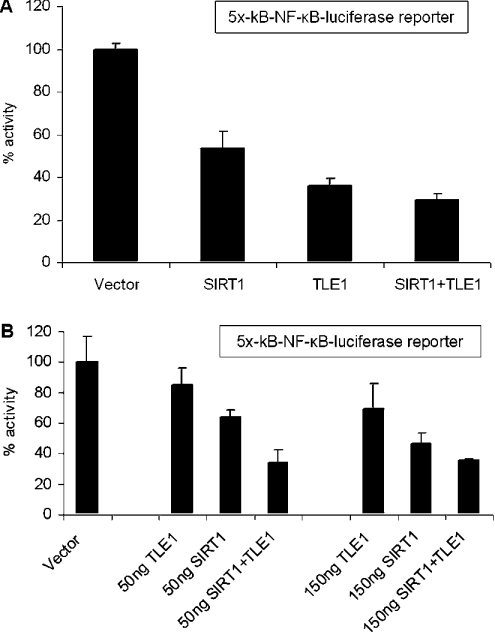
Both Sirt1 and TLE1 have been shown to regulate transcription mediated by NF-κB [14,16]. To examine the effect of Sirt1 and TLE1 on NF-κB-mediated gene expression, a co-transfection assay was performed using a luciferase reporter plasmid (5×-κB-TATA-Luc) containing five tandem repeats of the NF-κB DNA-binding sequence upstream of a TATA box. As shown in Figure 2(A), overexpression of Sirt1 and TLE1 both inhibited NF-κB-mediated luciferase reporter expression in HeLa cells, and the inhibitory effect was slightly enhanced when Sirt1 and TLE1 were both overexpressed.
Figure 2. Sirt1 and TLE1 repress NF-κB-dependent luciferase gene expression in HeLa cells.
(A) The results are presented as percentage luciferase activities. HeLa cells were transfected with 50 ng of 5×-κB-TATA-Luc reporter plasmid and 500 ng of the indicated expression plasmids. pCMV-3B, the backbone vector, was included so that all transfections had equivalent amounts of expression plasmids. Renilla luciferase was used as the internal transfection control. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase assay. (B) NF-κB-dependent luciferase gene repression mediated by Sirt1 and TLE1 is enhanced in the presence of both Sirt1 and TLE1 together. The results are presented as percentage luciferase activities. Cells were transfected with 50 ng of 5×-κB-TATA-Luc reporter plasmid and 0, 50 and 150 ng of the indicated expression plasmids. pCMV-3B, the backbone vector, was included so that all transfections had equivalent amounts of expression plasmids. Renilla luciferase was used as the internal transfection control. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase assay.
To investigate further the ability of the Sirt1–TLE1 interaction to inhibit NF-κB-mediated transcription, a dose–response analysis was performed using the NF-κB-dependent luciferase reporter (Figure 2B). At lower doses (50 and 150 ng) of Sirt1 and TLE1 expression plasmids, repression of NF-κB activity was increased by approx. 30–40% by co-expression compared with the repression mediated by Sirt1 and TLE1 individually. However, at the higher dose (500 ng) of expression plasmids, the difference between level of repression mediated by Sirt1 and TLE1 individually compared with when both the proteins were overexpressed was reduced. This result suggests that co-expression of Sirt1 and TLE1 together, at least at lower levels of expression, can mediate repression of NF-κB activity more efficiently than for either factor alone.
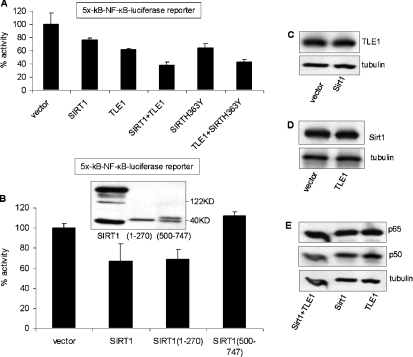
Deacetylase activity is not required for Sirt1-mediated repression of NF-κB
It has been reported that Sirt1 represses NF-κB activity by decetylating its p65 subunit, reducing its ability to transactivate [14]. Interestingly, the catalytic mutant of Sirt1, Sirt1-H363Y, was able to repress NF-κB activity in a co-transfection assay, similarly to wild-type Sirt1 (Figure 3A). In addition the inhibitory effect of the catalytic mutant was enhanced by co-transfection with TLE1. Given that the effect of Sirt1 on NF-κB transcriptional activity is independent of Sirt1 catalytic activity, we examined which region of Sirt1 was important for conferring the NF-κB inhibitory effect using expression plasmids for the N- (amino acids 1–270) and C- (amino acids 500–747) terminal fragments of Sirt1. As shown in Figure 3(B), overexpression of the N-terminal region of Sirt1 repressed NF-κB activity similarly to full-length Sirt1, whereas the C-terminal fragment of Sirt1 had no effect. As a further control, we examined the levels of expression of the exogenous Sirt1 and TLE1 as well as the endogenous NF-kB subunits p65 and p50. As shown in Figures 3(C) and 3(D), expression of Sirt1 did not affect the levels of expression of TLE1, and TLE1 did not affect expression of Sirt1. Moreover, there was no effect of TLE1 and Sirt1 expression on the levels of the endogenous NF-κB subunits (Figure 3E).
Figure 3. Deacetylase activity of Sirt1 is not required for repressing NF-κB activity.
The catalytic mutant Sirt1-H363Y (A) and the N-terminal (amino acids 1–270) fragment of Sirt1 (B) repress NF-κB-dependent luciferase gene expression. The results are presented as percentage luciferase activities. Cells were transfected with 50 ng of 5×-κB-TATA-Luc reporter plasmid and 500 ng of the indicated expression plasmids. pCMV-3B, the backbone vector, was included so that all transfections had equivalent amounts of expression plasmids. Renilla luciferase was used as the internal transfection control. At 24 h post-transfection cells were induced with TNFα for 3 h and harvested for the luciferase assay. Expression of N- and C-terminal fragments of Sirt1 is shown in the Western blot of the insert of (B). (C–E) Overexpression of Sirt1 and TLE1 individually or together does not affect exogenous protein expression or the endogenous NF-κB proteins p65 and p50. HeLa cells were transfected with Sirt1 or its backbone vector (C), TLE1 or its backbone vector (D) or TLE1 or Sirt1 or both (E). At 24 h post-transfection, the levels of TLE1, Sirt1 and p65–p50 were detected by Western blotting. An anti-tubulin antibody was used as a loading control.
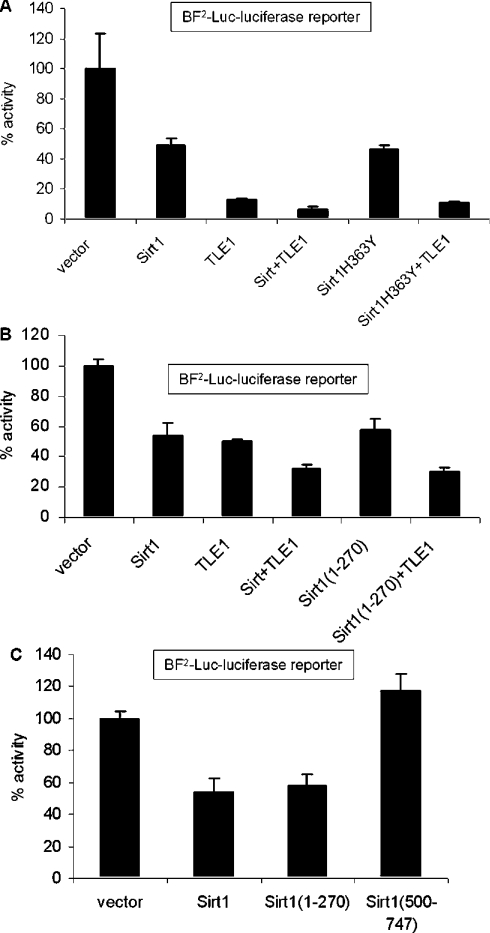
To determine whether Sirt1 and TLE1 also regulate transcription from non-synthetic NF-κB-binding sites, we used a natural NF-κB-responsive luciferase reporter, BF2-Luc, containing NF-κB sites from the IL-8 (interleukin 8) promoter. As shown in Figure 4, expression of either Sirt1 or TLE1 alone was also able to repress transcription from the NF-κB sites in the IL-8 promoter (Figure 4A), similarly to the synthetic NF-κB-binding site reporter. Furthermore, co-expression of Sirt1 and TLE1 together repressed NF-κB activity even more efficiently. Also, the catalytic mutant of Sirt1, Sirt1-H363Y (Figure 4A), and the N-terminus of Sirt1 (Figure 4B), which does not have the catalytic domain, were able to repress NF-κB activity similarly to the full-length Sirt1. This repression was enhanced further when co-expressed with TLE1. We have again shown using the IL-8 BF2-Luc reporter that, although the N-terminus of Sirt1 (amino acids 1–270) was able to repress NF-κB activity, no repression was observed with the C-terminus (amino acids 500–747) (Figure 4C).
Figure 4. Regulation of the NF-κB sites in the IL-8 promoter by Sirt1 and TLE1.
Cells were transfected with 50 ng of IL-8 promoter-based NF-κB-responsive BF2-Luc reporter plasmid and 500 ng of the indicated expression plasmids. pCMV-3B, the backbone vector, was included so that all transfections had equivalent amounts of expression plasmids. Renilla luciferase was used as the internal transfection control. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase assay. The results are presented as percentage luciferase activities.
Sirt1 and TLE1 are both required for inhibiting NF-κB activity
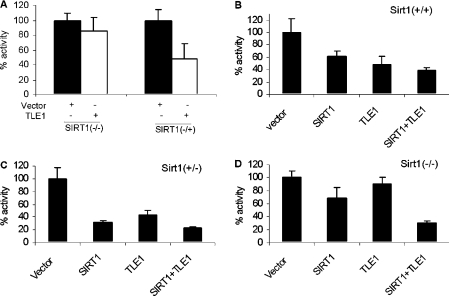
To determine whether Sirt1 is required for the inhibitory effect of TLE1 on NF-κB activity, the co-transfection assays with the NF-κB reporter were performed in MEFs that were either heterozygous null (−/+) or homozygous null (−/−) for Sirt1 [17]. As shown in Figure 5(A), TLE1-mediated repression of NF-κB transcriptional activity was significantly abrogated in Sirt1−/− MEFs even though TLE1 could repress NF-κB activity by 60% in Sirt1−/+ MEFs. Furthermore, as demonstrated in Figures 5(B), 5(C) and 5(D), the pattern of repression of NF-κB activity mediated by Sirt1 and TLE1 was restored in Sirt1−/− MEFs by transfection with the Sirt1 expression plasmid.
Figure 5. TLE1 is unable to repress NF-κB-dependent gene expression in MEFs that are null for Sirt1.
(A) MEFs that are homozygous Sirt1-null (−/−) or heterozygous SIRT1-null (−/+) were transfected with 50 ng 5×-κB-TATA-Luc reporter plasmid and 500 ng of the indicated expression plasmids. pCMV-3B was used as the vector control. Renilla luciferase was used as the internal control for transfection. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase assay. (B–D) In the presence of exogenously expressed Sirt1, TLE1 is able to repress NF-κB-dependent gene expression in Sirt1-null MEFs. MEFs that are wild-type (B), heterozygous Sirt1-null (C) or homozygous Sirt1-null (D) were transfected with 50 ng of 5×-κB-TATA-Luc reporter plasmid and 500 ng of the indicated expression plasmids. Renilla luciferase was used as the internal control for transfection. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase assay. The results are presented as percentage luciferase activities.
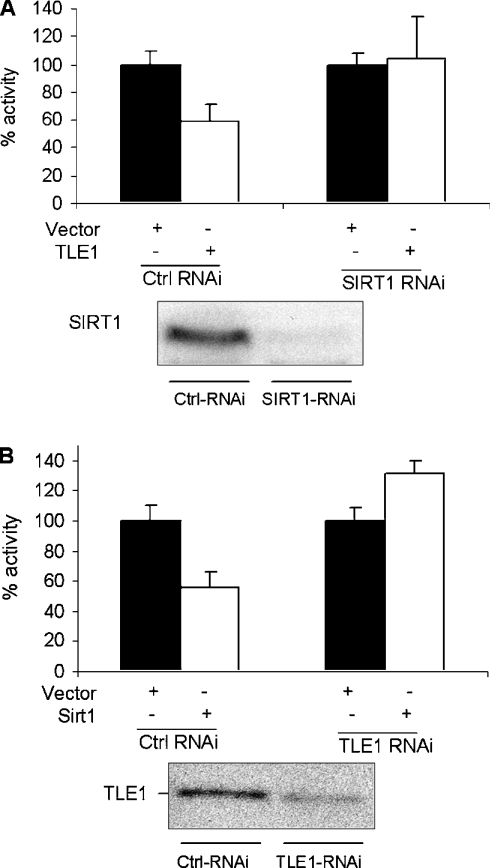
The results above suggest that the presence of Sirt1 is required for the inhibition of NF-κB by TLE1. To determine whether TLE1 was required for the suppression of NF-κB activity by Sirt1, HeLa cell populations that stably express siRNA to TLE1 or Sirt1 were established by retroviral transduction and selection. As shown in Figure 6, Sirt1 was unable to repress NF-κB in cells with reduced levels of TLE1. Conversely, in cells with reduced Sirt1 levels, TLE1 was unable to mediate repression of NF-κB activity, similarly to results from transfection of the Sirt1−/− MEFs. Taken together, our data suggest that an interaction between Sirt1 and TLE1 is important for the repression of NF-κB activity.
Figure 6. Sirt1 and TLE1 are both required for inhibiting NF-κB activity in HeLa cells.
The results are presented as percentage luciferase activities in HeLa cells that were depleted of either Sirt1 (A) or TLE1 (B) using siRNA. The cells were transfected with 50 ng of 5×-κB-TATA-Luc reporter plasmid and 500 ng of the indicated expression plasmids. Renilla luciferase was used as the internal transfection control. At 24 h post-transfection, cells were induced with TNFα for 3 h and harvested for luciferase analysis. Ctrl, control.
DISCUSSION
Sirt1 is able to repress transcription by associating with several transcription factors such as the bHLH repressor proteins HES1, HEY2 [1] and CTIP2 [2]. It also plays a direct role in repressive chromatin formation by interacting with the histone proteins H1, H3 and H4 [18]. Although yeast Sir2 is known to deacetylate histone proteins, its human homologue, Sirt1, has been also shown to deacetylate non-histone substrates such as p53, FOXO3/4, PCAF, MyoD, Ku70, PPARγ and PGC1α [5,7–9,11,19,20]. Furthermore, Sirt1 has been reported to inhibit NF-κB transcriptional activity by binding to and deacetylating p65/RelA [14,21]. Recently, Gao et al. [22] showed that the breast-cancer-associated protein BCA3 plays a role in recruiting Sirt1 to NF-κB. Furthermore, it was shown that NEDDylation of BCA3 is critical for its association with Sirt1 and hence for Sirt1-mediated transcriptional regulation of NF-κB. The results reported in the present paper and from other observations (J.V. Spencer, H.G. Ghosh and P.D. Robbins, unpublished work) from our group strongly suggest that Sirt1 is able to regulate NF-κB activity through a deacetylase-independent mechanism. Overexpression of both wild-type Sirt1 as well as a catalytic mutant of Sirt1, Sirt1-H363Y, repress a NF-κB-dependent promoter–luciferase reporter in HeLa cells. Moreover, the N-terminal fragment of Sirt1, which does not have any catalytic activity, can repress NF-κB activity. Also, repression mediated by Sirt1 was not abrogated by nicotinamide, an inhibitor of Sirt1 deacetylase activity (J.V. Spencer, H.G. Ghosh and P.D. Robbins, unpublished work).
In the present study, we have used a yeast two-hybrid screen to identify TLE1, a known repressor of NF-κB activity, as a Sirt1-interacting factor. The association between Sirt1 and TLE1 was confirmed further by co-immunoprecipitation of both endogenous and exogenous Sirt1 and TLE1. Moreover, we have shown that endogenous Sirt1 is required for TLE1-mediated repression of NF-κB activity, and, conversely, endogenous TLE1 is required for repression of NF-κB activity mediated by Sirt1. Taken together, our results suggest that Sirt1 is able to regulate NF-κB activity through a deacetylase-independent mechanism and requires TLE1.
TLE1 is the mammalian homologue of the Drosophila Groucho protein, a member of a family of transcriptional co-repressors. These co-repressors lack a DBD and are recruited to DNA through their interaction with DNA-binding proteins. Earlier studies on the Drosophila homologue of NF-κB, Dorsal, showed that it acts as both a transcriptional activator and a repressor depending upon its interaction with other transcriptional co-activators or co-repressors [23]. For example, Groucho binds directly to Dorsal, and converts it from a transcriptional activator into a repressor [15]. The mammalian Groucho homologues TLE1 and the AES also interact directly with mammalian NF-κB to repress its transcriptional activity [16]. However, the exact mechanism by which TLE1 associating with p65 leads to repression of NF-κB activity remains to be elucidated. In Drosophila, the recruitment of Groucho is known to be necessary, but not sufficient, for Dorsal-mediated repression and requires formation of a multiprotein DNA-binding complex consisting of Dorsal, Groucho and additional proteins such as cut and dead-ringer [24]. Hence, it is likely that TLE1 may require additional proteins such as Sirt1 to mediate the repression of NF-κB activity.
Interestingly, TLE1 and Sirt1 share certain biological properties. For example, Sirt1 and TLE1 are involved in mediating transcriptional repression by facilitating heterochromatin formation, TLE1 by virtue of it being a transcriptional co-repressor and Sirt1 by virtue of it being the histone deacetylase HDAC3. Both Sirt1 and TLE1 play a direct role in formation of repressive chromatin by associating with histone H3 [18,25]. Also, both of these proteins lack a DBD and are recruited to DNA by virtue of their interaction with various DNA-binding proteins and histone proteins. Furthermore, both of these proteins have been reported to associate with the RelA/p65 subunit of NF-κB, thereby repressing NF-κB-mediated gene transcription through a not yet clearly identified mechanism.
Our data demonstrate that neither Sirt1 nor TLE1 is able to repress NF-κB activity in the absence of the other. This suggests that the interaction between these proteins may play a critical role in repressing NF-κB activity. AES binds human p65 in the vicinity of its transactivation domain [16]. Thus it is possible that TLE1 recruits Sirt1 to the transactivation domain of p65 to deacetylate Lys310, mediating repression of NF-κB activity. However, as described above, our data also suggest that the catalytic activity of Sirt1 may not be critical for repression of NF-κB activity. Thus, alternatively, Sirt1 may play a role in recruiting the transcriptional co-repressor TLE1 to the p65 subunit of NF-κB, converting it from a transcriptional activator into a repressor by associating with histone proteins or other members of the transcriptional machinery. A recent study [10] demonstrated that the catalytic activity of Sirt1 may not be responsible for all of the functions of Sirt1. Using a new inhibitor molecule for Sirt1, EX-527, it was demonstrated that, even though inhibition of Sirt1 leads to increased p53 acetylation, it does not alter cell survival in response to DNA damage. This report [10] is in contrast with previous reports that suggested Sirt1 increases cellular resistance to DNA damage by deacetylating p53 and inhibiting p53-mediated apoptosis [9].
Even though the exact mechanism through which the interaction between Sirt1 and TLE1 help mediate repression of NF-κB activity is currently unclear, our results demonstrate, for the first time, that the mammalian Groucho protein TLE1 can associate with the sirtuin family protein Sirt1 to convert the transcriptional activator NF-κB into a transcriptional repressor. This result suggests that other transcriptional factors repressed by TLE1 may also be regulated by Sirt1 and, conversely, factors regulated by Sirt1 may also be regulated by TLE1.
Acknowledgments
We would like to thank Dr Roy Frye (University of Pittsburgh, Pittsburgh, PA, U.S.A.), Dr Marty Mayo (University of Virginia, Charlottesville, VA, U.S.A.) and Dr Robert Weinberg (Whitehead Institute of Biomedical Research, Cambridge, MA, U.S.A.) for providing reagents, and Dr Khaja K. Rehman and Dr Nicole Bianco for critical reading of the manuscript. This work was supported by DOD (Department of Defense) grants 17-03-1-0488 and 17-03-0142 to P. D. R.
References
- 1.Takata T., Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 2.Senawong T., Peterson V. J., Avram D., Shepherd D. M., Frye R. A., Minucci S., Leid M. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muth V., Nadaud S., Grummt I., Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerobo T. A., Miller C., Frye R., Ploegh H., Kessler B. M., Sinclair D. A. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 5.Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 6.Bouras T., Fu M., Sauve A. A., Wang F., Quong A. A., Perkins N. D., Hay R. T., Gu W., Pestell R. G. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 7.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 9.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 10.Solomon J. M., Pasupuleti R., Xu L., McDonagh T., Curtis R., Distefano P. S., Huber L. J. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Horst A., Tertoolen L. G., de Vries-Smits L. M., Frye R. A., Medema R. H., Burgering B. M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J. Biol. Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 12.Kamel C. Sirt1 fails to affect p53-mediated biological functions. Aging Cell. 2006;5:81–88. doi: 10.1111/j.1474-9726.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen L. F., Greene W. C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 14.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubnicoff T., Valentine S. A., Chen G., Shi T., Lengyel J. A., Paroush Z., Courey A. J. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tetsuka T., Uranishi H., Imai H., Ono T., Sonta S., Takahashi N., Asamitsu K., Okamoto T. Inhibition of nuclear factor-κB-mediated transcription by association with the amino-terminal enhancer of split, a Groucho-related protein lacking WD40 repeats. J. Biol. Chem. 2000;275:4383–4390. doi: 10.1074/jbc.275.6.4383. [DOI] [PubMed] [Google Scholar]
- 17.McBurney M. W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J. R., Lansdorp P. M., Lemieux M. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 20.Bereshchenko O. R., Gu W., Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 21.Chen J., Zhou Y., Mueller-Steiner S., Chen L. F., Kwon H., Yi S., Mucke L., Gan L. SIRT1 protects against microglia-dependent amyloid-β toxicity through inhibiting NF-κB signaling. J. Biol. Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 22.Gao F., Cheng J., Shi T., Yeh E. T. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFκB-dependent transcription. Nat. Cell. Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 23.Courey A. J., Huang J. D. The establishment and interpretation of transcription factor gradients in the Drosophila embryo. Biochem. Biophys. Acta. 1995;1261:1–18. doi: 10.1016/0167-4781(94)00234-t. [DOI] [PubMed] [Google Scholar]
- 24.Valentine S. A., Chen G., Shandala T., Fernandez J., Mische S., Saint R., Courey A. J. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol. Cell. Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palaparti A., Baratz A., Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 26.Lin S. Y., Elledge S. J. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 27.Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]