Abstract
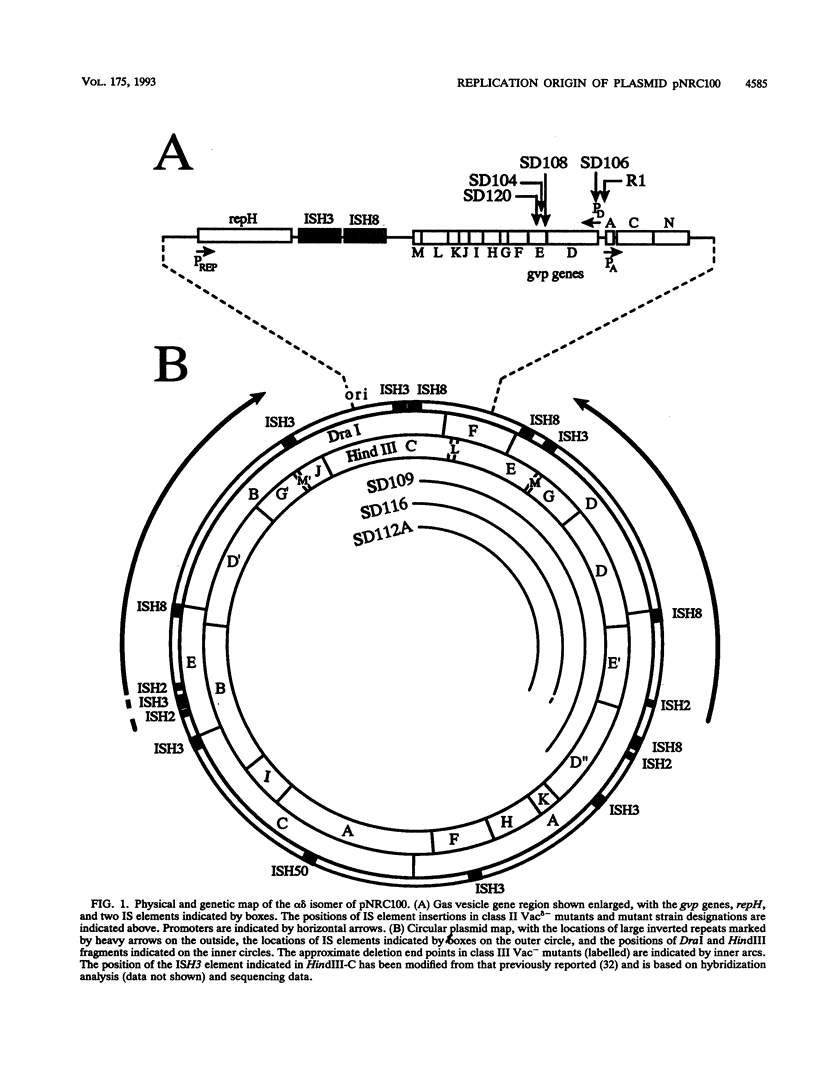
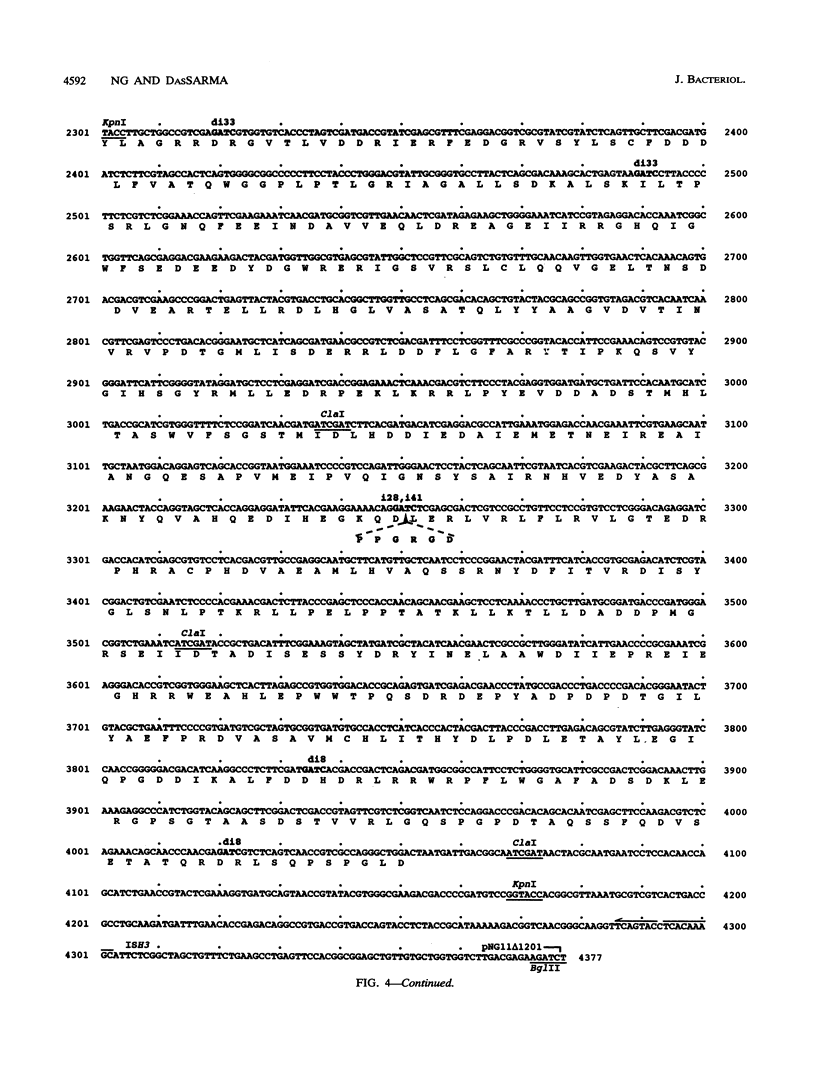
We have identified the replication origin of pNRC100, a 200-kb plasmid of Halobacterium halobium, by assaying for replication ability of miniplasmids containing cloned fragments of pNRC100 and the mevinolin resistance selectable marker of Haloferax volcanii. First, we showed the replication ability of plasmid pNGHCMEV1, which contains the 19-kb HindIII-C fragment of pNRC100, by recovery of plasmid DNA from mevinolin-resistant transformants of H. halobium. The minimal replication origin of approximately 3.9 kb was defined by subcloning successively smaller regions of pNGHCMEV1 and assaying for plasmid replication in either H. halobium or H. volcanii. The same replication origin was also recovered after transformation of H. volcanii with a library of partial Sau3AI fragments of pNRC100. The nucleotide sequence of the minimal replication origin was determined and found to contain a long open reading frame, named repH, transcribed away from a highly A+T-rich region. The transcription start site was identified by primer extension analysis to be 17 to 18 nucleotides 5' to a putative repH start codon. The predicted product of the repH gene, an acidic protein with a molecular weight of 113,442, showed 24 to 27% identity with predicted gene products of H. volcanii plasmid pHV2 and H. halobium plasmid p phi HL, suggesting that each is involved in plasmid replication. One pNRC100 minireplicon, pNG11 delta 12, was analyzed by linker scanning mutagenesis, which showed the requirement of repH for replication. Restoration of the repH reading frame of one replication-defective pNG11 delta 12 derivative by introduction of a second small insertion resulted in reversion to replication proficiency. The replication ability of pNG11delta12 was lost when the entire A+T-rich region, about 550 bp long, was deleted but not when small insertions or deletions were introduced into this region. The presence of only 52 bp of the A+T-rich segment was sufficient to permit replication. The pNG11delta12 minireplicon was lost at high frequency from cells grown without mevinolin selection, suggesting that the plasmid partitioning locus of pNRC100 is absent in the minimal replication origin region. We discuss the possible roles of the repH gene and the A+T-rich region in replication of pNRC100.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S. J. Plasmid partition. Plasmid. 1988 Jul;20(1):1–9. doi: 10.1016/0147-619x(88)90001-7. [DOI] [PubMed] [Google Scholar]
- Blaseio U., Pfeifer F. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois R. L., Lam W. L., Cline S. W., Doolittle W. F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Cline S. W., Doolittle W. F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987 Mar;169(3):1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S., Damerval T., Jones J. G., Tandeau de Marsac N. A plasmid-encoded gas vesicle protein gene in a halophilic archaebacterium. Mol Microbiol. 1987 Nov;1(3):365–370. doi: 10.1111/j.1365-2958.1987.tb01943.x. [DOI] [PubMed] [Google Scholar]
- DasSarma S. Mechanisms of genetic variability in Halobacterium halobium: the purple membrane and gas vesicle mutations. Can J Microbiol. 1989 Jan;35(1):65–72. doi: 10.1139/m89-010. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassarma S., Halladay J. T., Jones J. G., Donovan J. W., Giannasca P. J., de Marsac N. T. High-frequency mutations in a plasmid-encoded gas vesicle gene in Halobacterium halobium. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6861–6865. doi: 10.1073/pnas.85.18.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp F., Grampp B., Stolt P., Palm P., Zillig W. The immunity-conferring plasmid p phi HL from the Halobacterium salinarium phage phi H: nucleotide sequence and transcription. Virology. 1992 Sep;190(1):45–54. doi: 10.1016/0042-6822(92)91191-v. [DOI] [PubMed] [Google Scholar]
- Hackett N. R., DasSarma S. Characterization of the small endogenous plasmid of Halobacterium strain SB3 and its use in transformation of H. halobium. Can J Microbiol. 1989 Jan;35(1):86–91. doi: 10.1139/m89-013. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Halladay J. T., Jones J. G., Lin F., MacDonald A. B., DasSarma S. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J Bacteriol. 1993 Feb;175(3):684–692. doi: 10.1128/jb.175.3.684-692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halladay J. T., Ng W. L., DasSarma S. Genetic transformation of a halophilic archaebacterium with a gas vesicle gene cluster restores its ability to float. Gene. 1992 Sep 21;119(1):131–136. doi: 10.1016/0378-1119(92)90078-4. [DOI] [PubMed] [Google Scholar]
- Hofman J. D., Schalkwyk L. C., Doolittle W. F. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium, Halobacterium volcanii. Nucleic Acids Res. 1986 Sep 11;14(17):6983–7000. doi: 10.1093/nar/14.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. G., Hackett N. R., Halladay J. T., Scothorn D. J., Yang C. F., Ng W. L., DasSarma S. Analysis of insertion mutants reveals two new genes in the pNRC100 gas vesicle gene cluster of Halobacterium halobium. Nucleic Acids Res. 1989 Oct 11;17(19):7785–7793. doi: 10.1093/nar/17.19.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. G., Young D. C., DasSarma S. Structure and organization of the gas vesicle gene cluster on the Halobacterium halobium plasmid pNRC100. Gene. 1991 Jun 15;102(1):117–122. doi: 10.1016/0378-1119(91)90549-q. [DOI] [PubMed] [Google Scholar]
- Krebs M. P., RajBhandary U. L., Khorana H. G. Nucleotide sequence of ISH11, a new Halobacterium halobium insertion element isolated from the plasmid pGRB1. Nucleic Acids Res. 1990 Nov 25;18(22):6699–6699. doi: 10.1093/nar/18.22.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W. L., Doolittle W. F. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Beaucage S. L., Cohen S. N. Role of DNA superhelicity in partitioning of the pSC101 plasmid. Cell. 1990 Jul 13;62(1):127–133. doi: 10.1016/0092-8674(90)90246-b. [DOI] [PubMed] [Google Scholar]
- Ng W. L., Kothakota S., DasSarma S. Structure of the gas vesicle plasmid in Halobacterium halobium: inversion isomers, inverted repeats, and insertion sequences. J Bacteriol. 1991 Mar;173(6):1958–1964. doi: 10.1128/jb.173.6.1958-1964.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Betlach M. Genome organization in Halobacterium halobium: a 70 kb island of more (AT) rich DNA in the chromosome. Mol Gen Genet. 1985;198(3):449–455. doi: 10.1007/BF00332938. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Blaseio U. Insertion elements and deletion formation in a halophilic archaebacterium. J Bacteriol. 1989 Sep;171(9):5135–5140. doi: 10.1128/jb.171.9.5135-5140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Ghahraman P. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. Mol Gen Genet. 1993 Apr;238(1-2):193–200. doi: 10.1007/BF00279547. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Genetic variability in Halobacterium halobium. J Bacteriol. 1981 Jan;145(1):375–381. doi: 10.1128/jb.145.1.375-381.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Doolittle W. F. Unusual physical organization of the Halobacterium genome. Nature. 1982 Feb 4;295(5848):384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Sapienza C., Rose M. R., Doolittle W. F. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature. 1982 Sep 9;299(5879):182–185. doi: 10.1038/299182a0. [DOI] [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Simsek M., DasSarma S., RajBhandary U. L., Khorana H. G. A transposable element from Halobacterium halobium which inactivates the bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7268–7272. doi: 10.1073/pnas.79.23.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G., Klotz G., Goebel W. A large plasmid from Halobacterium halobium carrying genetic information for gas vacuole formation. Plasmid. 1979 Jul;2(3):377–386. doi: 10.1016/0147-619x(79)90021-0. [DOI] [PubMed] [Google Scholar]
- Womble D. D., Rownd R. H. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic resistance plasmids. Microbiol Rev. 1988 Dec;52(4):433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. L., Doolittle W. F. Structure of the archaebacterial transposable element ISH50. Nucleic Acids Res. 1983 Jun 25;11(12):4195–4199. doi: 10.1093/nar/11.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]