Abstract
Genetic and RNA interference (RNAi) screens for life span regulatory genes have revealed that the daf-2 insulin-like signaling pathway plays a major role in Caenorhabditis elegans longevity. This pathway converges on the DAF-16 transcription factor and may regulate life span by controlling the expression of a large number of genes, including free-radical detoxifying genes, stress resistance genes, and pathogen resistance genes. We conducted a genome-wide RNAi screen to identify genes necessary for the extended life span of daf-2 mutants and identified ∼200 gene inactivations that shorten daf-2 life span. Some of these gene inactivations dramatically shorten daf-2 mutant life span but less dramatically shorten daf-2; daf-16 mutant or wild-type life span. Molecular and behavioral markers for normal aging and for extended life span in low insulin/IGF1 (insulin-like growth factor 1) signaling were assayed to distinguish accelerated aging from general sickness and to examine age-related phenotypes. Detailed demographic analysis, molecular markers of aging, and insulin signaling mutant test strains were used to filter progeric gene inactivations for specific acceleration of aging. Highly represented in the genes that mediate life span extension in the daf-2 mutant are components of endocytotic trafficking of membrane proteins to lysosomes. These gene inactivations disrupt the increased expression of the DAF-16 downstream gene superoxide dismutase sod-3 in a daf-2 mutant, suggesting trafficking between the insulin-like receptor and DAF-16. The activities of these genes may normally decline during aging.
[Keywords: Aging, endocytosis, insulin, daf-2, C. elegans, RNAi]
The daf-2 signaling pathway is part of an endocrine system that regulates longevity, metabolism, and development in Caenorhabditis elegans and is homologous to the mammalian insulin and insulin-like growth factor 1 (IGF1) signaling pathway (Finch and Ruvkun 2001). daf-2 encodes the worm homolog of the insulin/IGF-I receptor. Signaling from DAF-2 is mediated through the AGE-1 phosphatidylinositol 3-kinase (PI3K), PDK-1, and AKT-1/2 kinases to antagonize DAF-16, orthologous to human FOXO, a forkhead transcription factor. The function of this pathway in mediating longevity and metabolism is conserved in C. elegans, Drosophila, and mammals (Finch and Ruvkun 2001; Garofalo 2002; Nakae et al. 2002; Bluher et al. 2003; Holzenberger et al. 2003).
Decreased DAF-2 signaling extends C. elegans’ life span up to threefold and causes increased fat storage and dauer arrest (Wadsworth and Riddle 1989; Kenyon et al. 1993; Kimura et al. 1997). The daf-2 insulin/IGF1 pathway also regulates the expression of free-radical detoxifying enzymes, consistent with free-radical theories of aging and the involvement of mitochondria and reactive oxygen species (ROS) in aging (Larsen 1993; Vanfleteren and De Vreese 1995; Honda and Honda 1999). These phenotypes are suppressed by daf-16 loss-of-function mutations, suggesting that daf-16 is negatively regulated by the daf-2 pathway and is the major downstream mediator of genes that extend life span (Lin et al. 1997; Ogg et al. 1997). The DNA-binding site of DAF-16 is present in many C. elegans genes (Furuyama et al. 2000), but only a few have been shown to be daf-2 responsive and fewer still have been shown to mediate the increase in longevity induced by low daf-2 signaling (Lee et al. 2003). Gene microarray analysis of daf-2 mutants have revealed thousands of transcripts that change in low daf-2 signaling but does not detect directly regulated genes (McElwee et al. 2003; Murphy et al. 2003; Golden and Melov 2004). daf-2 signaling from both the nervous system and intestine regulates life span (Apfeld and Kenyon 1998; Wolkow et al. 2000; Libina et al. 2003). While DAF-16 and more indirect transcriptional cascades are likely to mediate the process, the molecular mechanism of life span regulation remains unknown. Using RNA interference (RNAi) to screen for defects in daf-2 pathway-mediated longevity enhancement, we sought to identify the comprehensive genetic network necessary for long life span induction in a daf-2 insulin/IGF1 signaling- deficient mutant.
Results
daf-2 mutant animals were screened for increased mortality using an RNAi library containing 15,718 C. elegans genes to inactivate each of these genes. Escherichia coli strains expressing double-stranded RNAs (dsRNAs) derived from each of these C. elegans genes were fed to daf-2(e1370) mutant animals, allowing systematic inactivation of C. elegans genes by feeding dsRNA. Populations of daf-2 mutant animals synchronized at the first larval stage (L1) were placed on each E. coli strain expressing one particular dsRNA, and survival was measured prior to the normal time when daf-2 mutant populations start to die exponentially due to advancing age. daf-2 animals treated with E. coli expressing control dsRNA live up to 40 d at 25°C. daf-2 animals exposed to daf-16 dsRNA die before the 23rd day of adulthood. Feeding the dsRNA from the L1 stage allowed time for RNAi to become effective by the adult stage, when daf-2 activity is essential to extend life span (Dillin et al. 2002). Of the 15,718 gene inactivations tested, 1058 caused obvious developmental defects, confounding the study of shorter-than-normal life span (e.g., larval lethality, ruptured, blistered, and excessively small animals), as mortality may be due to sickness and not premature aging (data not shown). These genes may function in life span regulation but are more difficult to interpret and are not further characterized here. Of the remaining 14,660 gene inactivations without developmental pleiotropies, 500 decrease the life span of daf-2(e1370) mutant animals (3.4% of genes screened). The gene inactivations were primarily classified based on the magnitude of the decrease in daf-2 life span: One-hundred-twenty-seven gene inactivations produced >80% mortality at the time of scoring, compared with almost no mortality in the daf-2 control at that time.
The 500 positive RNAi clones from the primary screen were re-examined in a more detailed longitudinal life span analysis that recorded the survival curves of replicate populations of animals. For each gene inactivation, a cohort of age-synchronized daf-2 animals was divided into subpopulations. At each time point, the survival proportion within a different subpopulation of ∼25 animals was recorded in two or more independent experiments. Thus, the mortality measured in one subpopulation at an early point in life does not influence mortality in the second subpopulation scored at a later date. Additionally, scoring independent subpopulations normalizes the relative number of animals examined at each point in time and avoids the variability found late in the life of single populations as the number of surviving animals approaches zero. Since the mortality observed in the subpopulations occur independently, a more accurate measure of the mortality rate can be measured. Of the 500 positives, 159 gene inactivations caused a significantly shorter life span than control daf-2 animals, and this decrease in longevity was reproducible in multiple independent populations. The other 341 clones failed to significantly shorten the life span of daf-2 mutant animals in the longitudinal analysis or displayed developmental defects. Importantly, of those clones that failed to retest, only 10 were considered as “strong” positives in the primary screen, strengthening the confidence of life span observations made in the primary screen (Supplementary Table S1).
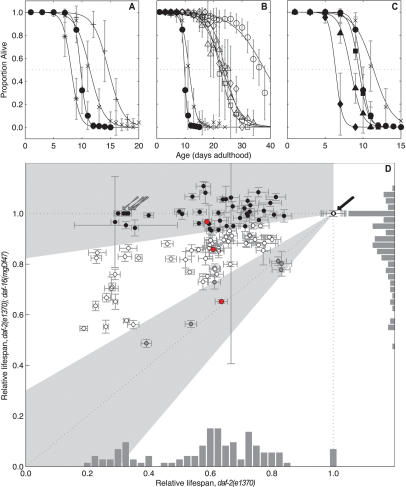
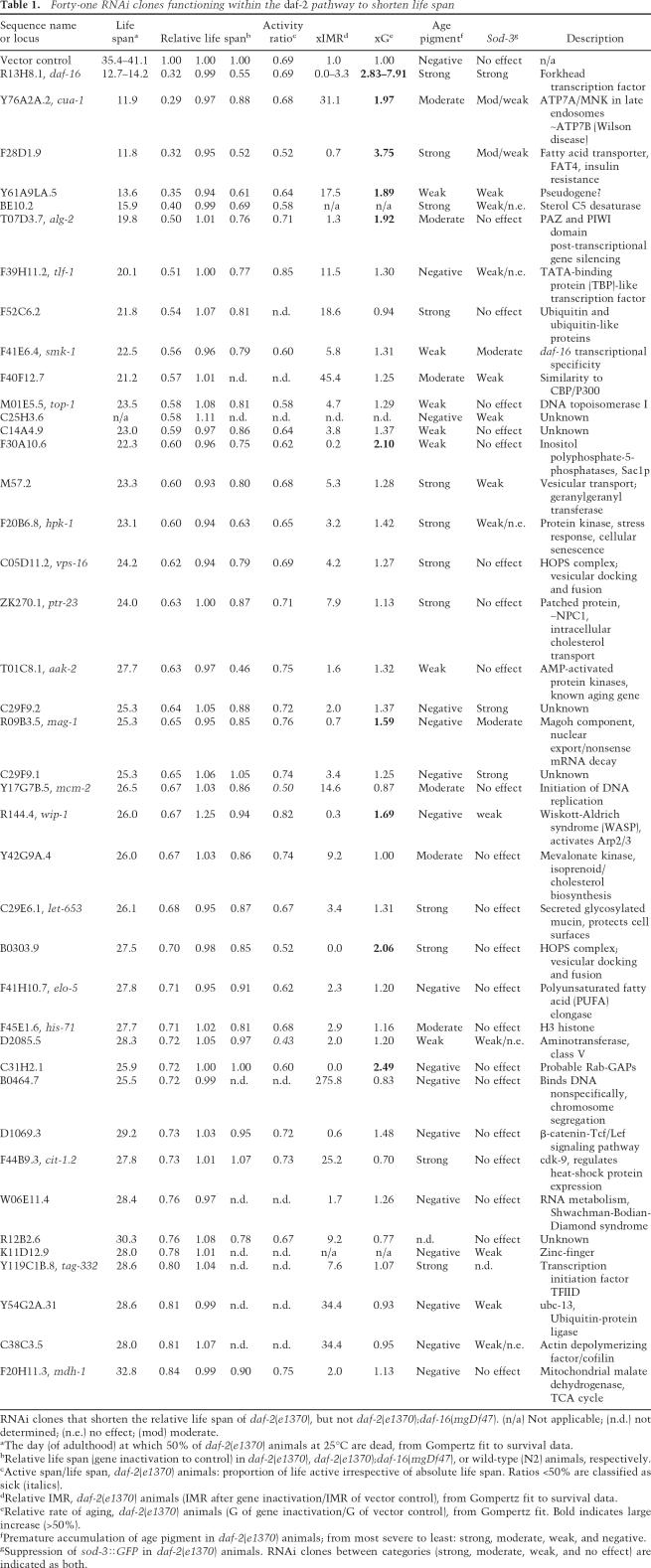
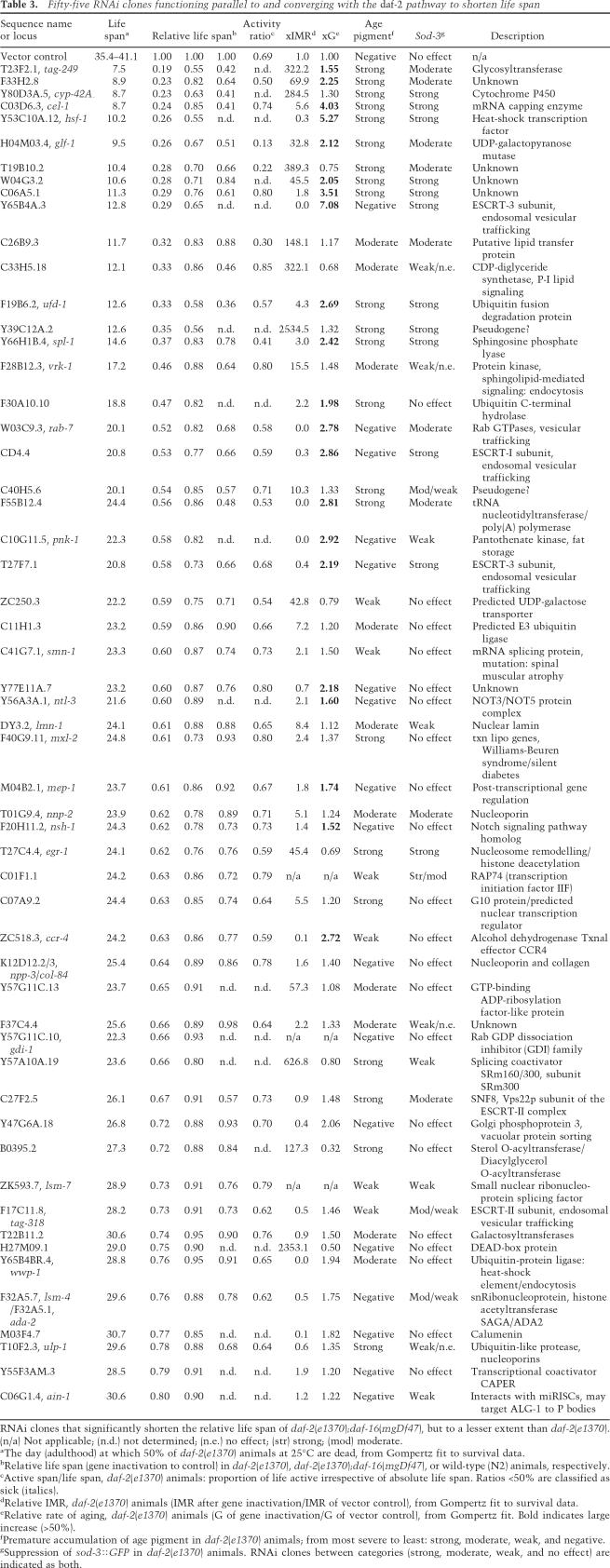
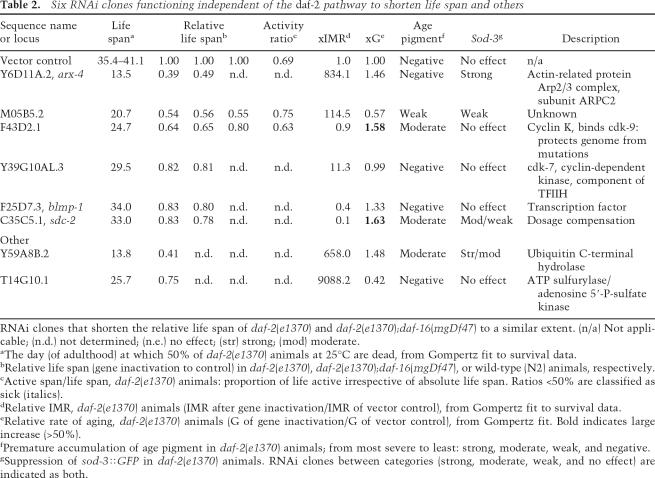
A key classification of the longevity genes was whether the gene inactivations shortened life span specifically in long-lived daf-2 mutant animals or affected life span in other genetic backgrounds as well. To test whether a particular gene inactivation is specific to the daf-2 pathway, we classified gene inactivations based on whether they shortened the life span of daf-2;daf-16 double-mutant animals, which decouple the insulin/IGF1 pathway. We also examined the impact of each gene inactivation on wild-type life span (Fig. 1A; summarized in Tables 1–3; Supplementary Table S2). Forty-one gene inactivations functioned specifically within the daf-2 pathway to shorten life span, not decreasing the life span of daf-2;daf-16 animals (Fig. 1D, black circles; Table 1; Supplementary Fig. S1; Supplementary Table S2). Six gene inactivations shortened life span in both daf-2 and daf-2; daf-16 strains; these genes regulate life span independently of insulin/IGF1 signaling (Fig. 1D, gray circles; Table 2; Supplementary Fig. S1; Supplementary Table S2). Fifty-seven gene inactivations (corresponding to 55 RNAi clones) more dramatically shortened the life span of daf-2 animals compared with daf-2;daf-16, but still shortened the life span of daf-2;daf-16 animals, suggesting that they function in a parallel/converging pathway to insulin/IGF1 signaling (Fig. 1D, white circles; Table 3; Supplementary Fig. S1; Supplementary Table S2). To illustrate this, RNAi against either C14A4.9, mep-1 (M04B2.1), or F43D2.1 in daf-2(e1370) each shortens life span ∼35%–40% (median life span of 22.4, 23.2, and 24.2 d; Fig. 1B, white squares, triangles, and diamonds, respectively), but these dsRNAs differentially shorten the life span of daf-2;daf-16 mutant animals (Fig. 1C,D, red circles).
Figure 1.
Genetic specificity as a classification tool for RNAi clones shortening the life span of daf-2 mutant animals. (A) Loss of daf-16 shortens N2 life span. Using wild type as a reference strain to define daf-2 pathway specificity would classify daf-16 as outside the daf-2 pathway. Thus, wild-type animals cannot be used to categorize whether a particular gene inactivation functions in the daf-2 pathway. (+) N2 (wild type); control RNAi; (X) daf-2(e1370);daf-16 RNAi; (●) daf-2(e1370);daf-16(mgDf47); control RNAi; (*) N2;daf-16 RNAi. (B) Three RNAi clones all shorten the life span of daf-2(e1370) animals to a similar extent. (◦) daf-2(e1370); control RNAi; (◽) daf-2(e1370);C14A4.9 RNAi; (▵) daf-2(e1370);mep-1 RNAi; (◊) daf-2(e1370);F43D2.1 RNAi. (C) However, these same RNAi clones cause distinct changes in the life span of daf-2(e1370);daf-16(mgDf47) animals. A typical daf-2 pathway-specific gene inactivation shortens the life span of daf-2(e1370) animals but not daf-2(e1370);daf-16(mgDf47) double-mutant animals; e.g., daf-2(e1370);daf-16(mgDf47);C14A4.9 RNAi (◼). A typical gene inactivation functioning in a parallel/converging pathway to daf-2 significantly shortens the life span of daf-2(e1370);daf-16(mgDf47) animals, but to a lesser degree than in daf-2(e1370) animals; e.g., daf-2(e1370);daf-16(mgDf47);mep-1 RNAi (▴). A gene inactivation functioning completely independently from the daf-2 pathway shortens daf-2(e1370) and daf-2(e1370);daf-16(mgDf47) life span to the same degree; e.g., daf-2(e1370);daf-16(mgDf47);F43D2.1 RNAi (◆). (X) daf-2(e1370);daf-16 RNAi; (●) daf-2(e1370);daf-16(mgDf47);control RNAi. (D) Summary for each gene inactivation, comparing the induced life span on daf-2(e1370) (X-axis) versus daf-2(e1370);daf-16(mgDf47) (Y-axis). For each RNAi clone, the life span is relative to control RNAi in each strain. Pathway specificity: (●) Strictly daf-2; (◦) parallel/converging on daf-2; (●) independent of daf-2. RNAi clones in B and C are marked in red. Gray regions delineate the classes of pathway specificity. (Gray arrow) daf-16 RNAi; (black arrow) empty vector control RNAi.
Table 1.
Forty-one RNAi clones functioning within the daf-2 pathway to shorten life span
RNAi clones that significantly shorten the relative life span of daf-2(e1370);daf-16(mgDf47), but to a lesser extent than daf-2(e1370). (n/a) Not applicable; (n.d.) not determined; (n.e.) no effect; (str) strong; (mod) moderate.
aThe day (adulthood) at which 50% of daf-2(e1370) animals at 25°C are dead, from Gompertz fit to survival data.
bRelative life span (gene inactivation to control) in daf-2(e1370), daf-2(e1370);daf-16(mgDf47), or wild-type (N2) animals, respectively.
cActive span/life span, daf-2(e1370) animals: proportion of life active irrespective of absolute life span. Ratios <50% are classified as sick (italics).
dRelative IMR, daf-2(e1370) animals (IMR after gene inactivation/IMR of vector control), from Gompertz fit to survival data.
eRelative rate of aging, daf-2(e1370) animals (G of gene inactivation/G of vector control), from Gompertz fit. Bold indicates large increase (>50%).
fPremature accumulation of age pigment in daf-2(e1370) animals; from most severe to least: strong, moderate, weak, and negative.
gSuppression of sod-3∷GFP in daf-2(e1370) animals. RNAi clones between categories (strong, moderate, weak, and no effect) are indicated as both.
Table 3.
Fifty-five RNAi clones functioning parallel to and converging with the daf-2 pathway to shorten life span
RNAi clones that significantly shorten the relative life span of daf-2(e1370);daf-16(mgDf47), but to a lesser extent than daf-2(e1370). (n/a) Not applicable; (n.d.) not determined; (n.e.) no effect; (str) strong; (mod) moderate.
aThe day (adulthood) at which 50% of daf-2(e1370) animals at 25°C are dead, from Gompertz fit to survival data.
bRelative life span (gene inactivation to control) in daf-2(e1370), daf-2(e1370);daf-16(mgDf47), or wild-type (N2) animals, respectively.
cActive span/life span, daf-2(e1370) animals: proportion of life active irrespective of absolute life span. Ratios <50% are classified as sick (italics).
dRelative IMR, daf-2(e1370) animals (IMR after gene inactivation/IMR of vector control), from Gompertz fit to survival data.
eRelative rate of aging, daf-2(e1370) animals (G of gene inactivation/G of vector control), from Gompertz fit. Bold indicates large increase (>50%).
fPremature accumulation of age pigment in daf-2(e1370) animals; from most severe to least: strong, moderate, weak, and negative.
gSuppression of sod-3∷GFP in daf-2(e1370) animals. RNAi clones between categories (strong, moderate, weak, and no effect) are indicated as both.
Table 2.
Six RNAi clones functioning independent of the daf-2 pathway to shorten life span and others
RNAi clones that shorten the relative life span of daf-2(e1370) and daf-2(e1370);daf-16(mgDf47) to a similar extent. (n/a) Not applicable; (n.d.) not determined; (n.e.) no effect; (str) strong; (mod) moderate.
aThe day (of adulthood) at which 50% of daf-2(e1370) animals at 25°C are dead, from Gompertz fit to survival data.
bRelative life span (gene inactivation to control) in daf-2(e1370), daf-2(e1370);daf-16(mgDf47), or wild-type (N2) animals, respectively.
cActive span/life span, daf-2(e1370) animals: proportion of life active irrespective of absolute life span. Ratios <50% are classified as sick (italics).
dRelative IMR, daf-2(e1370) animals (IMR after gene inactivation/IMR of vector control), from Gompertz fit to survival data.
eRelative rate of aging, daf-2(e1370) animals (G of gene inactivation/G of vector control), from Gompertz fit. Bold indicates large increase (>50%).
fPremature accumulation of age pigment in daf-2(e1370) animals; from most severe to least: strong, moderate, weak, and negative.
gSuppression of sod-3∷GFP in daf-2(e1370) animals. RNAi clones between categories (strong, moderate, weak, and no effect) are indicated as both.
Many of these potential progeric genes also shortened wild-type life span. Twenty gene inactivations, such as the ubiquitin-like protein F52C6.2, shortened daf-2 life span to a much greater extent than either daf-2; daf-16 or N2 life span, and 27 gene inactivations caused little decrease (0%–20% decreased life span) of the relative life span of wild-type animals as well as daf-2; daf-16 double-mutant animals. Ten gene inactivations shortened both wild-type and daf-2; daf-16 life spans, but shortened the relative life span of daf-2 mutants to a greater extent. Like daf-16 RNAi, 19 clones shortened wild-type life span (>20% decreased life span) without decreasing daf-2; daf-16 life span (0%–20%). Overall, gene inactivations tended to shorten the relative life span of daf-2 mutant animals more than wild type, even though many gene inactivations shorten both wild-type and daf-2 life span (summarized in Supplementary Table S2; Supplementary Figs. S2, S3).
Gene inactivations decreasing daf-2 life span more than wild-type or daf-2;daf-16 life span are likely to be genes regulated by insulin/IGF1 signaling to in turn protect organisms from the ravages of aging. Those gene inactivations that decrease the life span of both daf-2 and wild-type animals could still constitute the components of life span regulation that are active in both normal and extended longevity life spans. It is probably significant that no gene inactivation affected wild-type or daf-2;daf-16 life spans more than daf-2 life span, showing that insulin/IGF1 regulation of life span cannot supersede other axes of longevity.
A persistent issue for any genetic change that leads to premature aging, or progeria, is whether it is not simply a general sickness that in turn causes early demise. Any screen to study aging by shortening life span must have a means to distinguish aging from sickness. We therefore assessed these progeric gene inactivations for whether they show molecular and demographic evidence of faster aging.
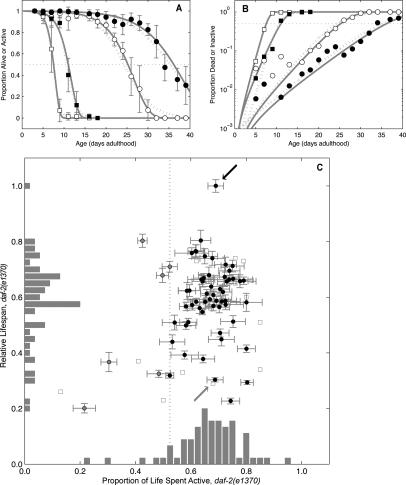
Degenerative changes occur in C. elegans that are correlative with eventual mortality. Specifically, as C. elegans age a transition from fast body movement to reduced motor activity occurs followed by a second transition where pharyngeal pumping ceases (Huang et al. 2004). Thus, a decrease in movement rate is one measure of aging. Adding water to a well and measuring thrashing rate, both daf-2(e1370) and wild-type synchronized subpopulations were simultaneously measured for life span and for a transition from activity to lethargy (defined here as less than one full movement back and forth per second). For instance, at the 26th day of adulthood, on average ∼90% of daf-2(e1370) animals are alive (Fig. 2A, black circles) but ∼40% actively thrash in water (Fig. 2A, white circles). Animals undergo an exponential decrease in movement as a function of age that can be observed by comparing movement data with life span plotted as the log(proportion dead or inactive) over time (Fig. 2B, cf. white and black circles and cf. white and black squares, respectively). This analysis revealed that the rates of mortality and movement decline are correlated: The aging coefficients (rate of aging determined from demographic analysis, discussed below) for daf-2(e1370) animals on control and daf-16 RNAi (0.18 ± 0.02 and 0.56 ± 0.05, respectively) differ by a factor of 3, yet the ratios of these coefficients (movement/mortality) are similar (1.47 ± 0.19 and 1.66 ± 0.16, respectively). Thus, decline in motor activity positively correlates with aging, and implies that testing whether changes in activity occur in short-lived animals is a means to distinguish accelerated aging from sickness.
Figure 2.
A shortened relative life span without changing the proportion of an animal’s life spent active distinguishes gene inactivations that accelerate aging. (A) High proportions of inactivity (white symbols) precede high proportions of mortality (black symbols). Shown are daf-2(e1370) animals treated with dsRNA to an empty vector control (circles) or to daf-16 (squares). Error bars are population-weighted standard deviations. Both Gompertz (solid gray line) and logit (dotted gray line) fits are shown. (B) Data and fits replotted with log(proportion) scale indicates that the rate of transition from active to lethargic movement and the mortality rate are positively correlated. (C) Clones with proportion active significantly <1. A vertical line indicates an arbitrary cutoff (lower 10% by clone rank) to identify sick clones (gray circles); e.g., those with proportion active <0.52. All other clones (black circles) are, by this definition, healthy. While clones are widely distributed by relative life span, they are clustered vertically near the empty vector control clone (black arrow) and daf-16 RNAi clone (gray arrow) in terms of proportion of life spent active. Error bars indicate mean ± SD. Clones with unquantified uncertainty in life span or activity ratio, derived as the quotient of mean active span to mean life span, are also shown (squares).
The transition from active to lethargic movement was measured longitudinally and compared with mortality to examine whether a particular gene inactivation altered the active span and activity fraction, the absolute number of days animals are active, and the fraction of life span in which animals were active, respectively (Fig. 2C; summarized in Tables 1–3; Supplementary Fig. S4; summarized in Supplementary Table S3). Control daf-2(e1370) animals have a median active span of 24.3 ± 0.9 (SD) days and median life span of 35.4 ± 0.6 (SD) days, or an activity fraction of 63.5%–74.5% (95% confidence interval or CI); daf-2(e1370) animals on daf-16 RNAi have a nearly identical activity fraction (63.0%–74.5%, 95% CI) despite a dramatically different active span (7.4 ± 0.3 SD) and life span (10.7 ± 0.2 SD). Based on these results, we classified any gene inactivations reducing the active portion of life span to less than ∼50% of control as sick, with the caveat that these gene inactivations may accelerate an early aspect of aging without drastically affecting late life mortality. Nine clones met this criteria and, of these, all but two had an initial mortality rate (IMR) increase by 14- to 389-fold relative to vector, and five had strong increases in the rate of aging (discussed below). Given that gene inactivations had already been filtered based on progeny production (see below), developmental pleiotropies, and, to some extent, age pigment production, it was not surprising that only a few gene inactivations shortened the activity ratio without an increased IMR. This suggests that gene inactivations that shorten life span can have varying impact on the mortality rate doubling time (MRDT), IMR, and the activity fraction. Furthermore, gene inactivations that shorten life span and cause lethargy throughout life promote sickness and rarely increase the rate of aging.
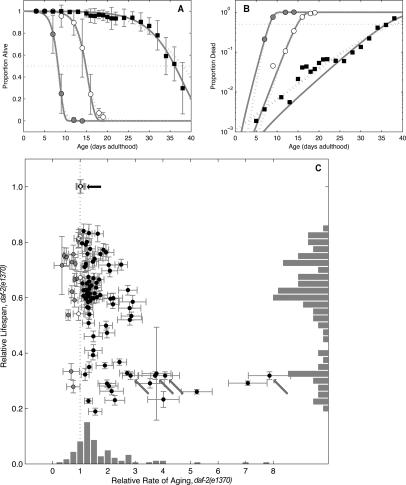
Many organisms, including C. elegans, have an exponential increase in the force of mortality with chronological age that can be approximated by the Gompertz model (for review, see Pletcher et al. 2000). Characterization of the mortality phenotype for each gene inactivation distinguishes whether life span is shortened due to an increased IMR or a decrease in the MRDT. Changes in the IMR indicate an increased risk of death throughout life or, more simply, a reduction in the period of time prior to the onset of aging. In the case of MRDT, a decrease implies an increase in the rate of aging. We calculated IMR, MRDT, and median age of death from a Gompertz model best fit to the survival data {Fig. 3A [proportion alive], B [log(proportion dead)]} for each gene inactivation that shortened the life span of daf-2(e1370) animals (summarized in Tables 1–3; Supplementary Table S4). Consistent with previous findings in C. elegans and flies (Johnson 1990; Gems and Riddle 2000; Tu et al. 2002), daf-2(e1370) controls (Fig. 3A,B, black squares) have a longer MRDT (3.25 d at 25°C) than wild-type animals (1.11 d at 25°C) (Fig. 3A,B, white circles) and a slightly lower IMR, implying that the rate of aging in daf-2(e1370) animals is lower than in wild-type C. elegans. In contrast, wild-type animals have a smaller MRDT (0.75 d at 25°C) when daf-16 is inactivated (Fig. 3A,B, gray circles), consistent with prior findings that daf-16 mutant animals are slightly progeric (summarized in Supplementary Table S5). Thus, an independent sampling of population mortality for each data point allows demographic analysis for each gene inactivation that shortens life span to determine whether there is an increase in the probability of dying (increased IMR) and/or an increase in the rate of aging (decreased MRDT) that, in conjunction with the other aforementioned assays, distinguishes sickness from accelerated aging.
Figure 3.
Decreased insulin signaling extends life span and decreases the rate of aging. In contrast, loss of daf-16 and many of the progeric gene inactivations described here shortens life span and increases the rate of aging. (A) Pooled mortality proportions for wild type (N2); empty vector control RNAi (◦), N2; daf-16 RNAi (●), and daf-2(e1370);vector control RNAi (◼). Error bars are population-weighted standard deviations. Both Gompertz (dark-gray line) and logit (light-gray line) fits are shown. (B) Data and fits replotted with log(proportion) scale. (C) Eighty-one gene inactivations had MRDT ratios significantly >1 (black circles), implying faster aging than empty vector control (black arrow). An additional 14 gene inactivations cause slower rates of aging (gray circles), implying that they were short-lived due to a higher probability of dying throughout life unrelated to aging (increased IMR). For four gene inactivations the rate of aging showed no significant difference (white circles) relative to empty vector control. Error bars indicate mean ± SD. Histograms show relative count of clones within each bin. Three data sets of daf-16 RNAi (gray arrows) were significant; these clones showed significant variation in MRDT ratio. The increasing variation reflects an increasing steepness in slope (i.e., a very high rate of aging) that is difficult to accurately measure, given that life span is being scored every other day in daf-2(e1370).
Seventy-seven gene inactivations that shorten daf-2 mutant life span produced a significant increase in the rate of daf-2 aging (Fig. 3C, black circles; summarized in Tables 1–3; Supplementary Fig. S5; summarized in Supplementary Table S4). Twelve had an IMR >30-fold higher than control, implying a pleiotropic phenotype. Including daf-16, 32 gene inactivations increased the rate of aging by >50% relative to daf-2(e1370) animals. Nine of these 32 gene inactivations function exclusively in the daf-2 pathway, while 20 function in a parallel/converging pathway. As the rate of aging increases, the steepness of the slope (related to the Gompertz aging coefficient, G) increases to a point where the frequency at which animals are scored for viability becomes limiting, which explains the variation in the rate of aging seen with daf-16 RNAi (in four data sets: fold G of 2.8, 3.7, 4.1, and 7.9) (Fig. 3C, gray arrows). Nevertheless, 11 gene inactivations produced an increased rate of aging on a par with the loss of daf-16: Y65B4A.3, hsf-1, cel-1, F28D1.9, C06A5.1, pnk-1, CD4.4, F55B12.4, rab-7, ccr-4, and ufd-1.
Aging C. elegans show intestinal autofluorescence partially as a result of lysosomal deposits of the pigment lipofuscin. Aging tissues accumulate this pigment in secondary lysosomes due to the oxidative degradation and autophagocytosis of cellular components (Klass 1977; Russell and Seppa 1987; Brunk and Terman 2002). daf-2 mutant animals show delayed onset of lipofuscin accumulation (Garigan et al. 2002), and loss of daf-16 causes earlier accumulation of lipofuscin in daf-2 animals (Gerstbrein et al. 2005). We tested whether gene inactivations that shortened life span also led to a premature accumulation of age pigment. Out of 39 strictly daf-2 pathway-specific clones, 17 gene inactivations caused strong or moderate acceleration in age pigment accumulation (Table 1). Similarly, of 66 gene inactivations that shorten both daf-2 and daf-2;daf-16 life span, 32 gene inactivations produced a strong/moderate premature accumulation of age pigment (Table 2). These data suggest that both classes of genes are truly progeric. To test whether premature accumulation of age pigment occurs in developmentally sick animals, we tested 11 gene inactivations that were scored as “pleotropic” due to developmental defects, yet were healthy enough to produce adult animals that could be assayed for age pigment accumulation. Out of the 11 pleiotropic gene inactivations, only one scored as strong or moderate premature accumulation of age pigment; gene inactivations that shorten life span via sickness do not score as progeric by this test. To test whether our genes that shorten life span were enriched for premature age pigment accumulation over what is expected by random, we compared these results to a pilot RNAi screen covering chromosome 4 for accelerated accumulation of age pigment in daf-2(1370), which revealed 79 positives out of 1392 gene inactivations tested (data not shown). Thus, gene inactivations functioning exclusively in the daf-2 pathway or in a parallel pathway converging with the daf-2 pathway were significantly enriched for an increase in the rate at which age pigment accumulates (P = 5E−23 and 2E−49, respectively).
Premature age pigment accumulation correlates with short life span, lethargy, and increased rate of aging. A large reduction of daf-2 median life span (<19 d) occurred with 25 gene inactivations, of which 22 also accelerated the accumulation of age pigment (88%). In contrast, of 79 gene inactivations causing a more modest decrease in median life span (>19 d), only 39% also caused a premature accumulation of age pigment. Additionally, of the nine gene inactivations that make animals “sick” by reducing the activity ratio (to <50%), eight had accelerated accumulation of age pigment. Of note, life span and activity ratio are not significantly correlated (P = 0.128) for large life span reductions (<50%). Thus lethargy also correlates with accumulation of age pigment, irrespective of the absolute reduction in life span, and implies that assessing premature age pigment accumulation alone is insufficient to distinguish sickness from accelerated aging. However, of 22 gene inactivations producing a twofold or greater increase in the rate of aging, 12 also prematurely accumulate age pigment. Interestingly, seven of those without premature age pigment accumulation are implicated in vesicular trafficking or protein sorting, which could directly inhibit the accumulation of age pigment and consequently be functionally equivalent to age pigment accumulation (discussed in more detail below). Thus, 80% (12 out of 15) of the gene inactivations with the greatest increase in the rate of aging also prematurely accumulate age pigment, and 86% of gene inactivations with greater than a twofold increase in the rate of aging (xG) also have a premature breakdown in vesicular trafficking to lysosomes or protein sorting (defined either by the accumulation of age pigment or by gene annotation). Collectively, these results demonstrate a correlation between age pigment accumulation and a shortened life span, lethargy, and an increased rate of aging. Whether age pigment accumulation causes aging or is just a symptom is a question that remains unresolved. However, our results suggest a common mechanism by which age pigment accumulation may cause aging. Our results show that disrupting endosomal protein sorting or blocking vesicular trafficking to the lysosome through RNAi increases the rate of aging. One consequence of blocking lysosomal trafficking is the improper recycling of membrane proteins (e.g., Papini et al. 1997; Vitelli et al. 1997). As age pigment accumulates in lysosomes, lysosomal function decreases (for reviews, see Terman and Brunk 2006; Terman et al. 2006). Collectively, this suggests that in normal aging the accumulation of age pigment ultimately leads to the breakdown of vesicular trafficking and autophagy, which in turn would result in improper recycling of membrane components—both of which cause accelerated aging.
The potential progeric gene inactivations were also screened for pleiotropies such as decreased fertility. Those gene inactivations that strongly affect brood size could either be generally unhealthy or, as in the case of daf-2 mutant animals, longevity may be coregulated with reproduction. In contrast, while daf-16(0) animals are progeric, with a shortened life span, increased stress sensitivity, and accelerated accumulation of age pigment, these animals have normal brood sizes. Thus, one way to separate gene inactivations that shorten life span is on the basis of whether they do so without drastically impairing production of progeny. Out of 185 positives, 55 dramatically decreased fecundity (<50% of a normal brood) (summarized in Supplementary Table S1). Filtering out small brood size producers eliminated ftt-2 (F52D10.3), a 14–3–3 protein previously shown to be involved in aging (Berdichevsky et al. 2006; Wang et al. 2006; Li et al. 2007). Additionally, accessing levels of age pigment in 47 of the clones producing decreased fecundity revealed 23% with premature age pigment accumulation, a significant enrichment (P = 8E−07) (data not shown). Thus, while filtering genes with reduced progeny eliminates some genes that may regulate aging, the remaining gene inactivations are less likely to have energetic issues or a subtle developmental defect and thus are more likely to regulate life span rather than just cause general unhealthiness.
To address whether the progeric gene inactivations regulate aging via daf-16, we looked for changes in the levels of a bona fide transcriptional target of DAF-16, sod-3 (Honda and Honda 1999). Decreased daf-2 signaling causes dephosphorylation and subsequent nuclear localization of DAF-16 (Lee et al. 2001). sod-3 contains two DAF-16 DNA-binding sites, and decreased insulin/IGF1 signaling results in the daf-16-dependent up-regulation of sod-3, which is easily visible in a sod-3∷GFP reporter. sod-3 is a manganese superoxide dismutase that functions to protect cells from oxidative stress and whose function may promote normal life span, albeit loss of sod-3 has not been shown to shorten daf-2 longevity. Each gene inactivation was tested in daf-2 mutant animals coexpressing a sod-3∷GFP reporter by following animals from early adulthood. Gene inactivations were classified on the basis of both the magnitude and time of sod-3∷GFP expression (early vs. late life span) to identify genes required for decreased insulin/IGF1 signaling to maintain the induction of sod-3 (summarized in Tables 1–3). Thirty-four gene inactivations suppressed the daf-2-dependent induction of sod-3 expression in nonneuronal cells. Gene inactivations that suppressed sod-3∷GFP expression included expected genes, such as hsf-1. Surprisingly, only six strictly daf-2 pathway-specific gene inactivations also suppressed sod-3 expression: C29F9.1, C29F9.2, smk-1, mag-1, F28D1.9, and cua-1. Only loss of sdc-2 or arx-4 suppresses sod-3 expression and genetically functions independently of the daf-2 pathway to shorten life span. The remaining 26 gene inactivations suppress sod-3 expression and function genetically in parallel to the daf-2 pathway to affect longevity, suggesting that their impact on daf-2 life span converges upstream of daf-16. SMK-1 is known to regulate DAF-16 transcriptional specificity and functions downstream from daf-16 nuclear localization (Wolff et al. 2006). Multiple genes involved in vesicular trafficking to lysosomes were required for sod-3 induction (discussed below). MAG-1 is the homolog of the mago nashi protein, which mediates mRNA splicing, nuclear export, and nonsense-mediated mRNA decay (Li et al. 2000), suggesting a link between mRNA fidelity and aging. Consistent with this possibility, even though the function of C29F9.1 and C29F0.2 in C. elegans is unknown, the former shares similarity with a Saccharomyces cerevisiae myosin-like nuclear envelope protein that functions in the nuclear retention of unspliced mRNAs and the latter shares similarity with a cytoplasmic nucleoporin required for polyadenylated RNA export. cel-1, an mRNA capping enzyme, and the nucleoporin npp-2 also suppress sod-3 expression when inactivated, further implying a possible link between aging and mRNA fidelity or somatic maintenance.
Many of the progeric genes identified in our screen are annotated to mediate vesicle sorting. Inactivation of these genes alters the morphology of GFP translational fusions to vesicular markers expressed in the intestine: RAB-5∷GFP labels early endosomes, RAB-7∷GFP labels early and late endosomes, RAB-10∷GFP labels endosomes and parts of the Golgi, and GES-1∷GFP labels the peroxisome. Each strain on control RNAi had punctuated GFP expression. In all cases, no change in the Golgi or peroxisome was observed (summarized in Supplementary Table S6). Not surprisingly, rab-7 RNAi suppressed the expression of RAB-7∷GFP but did not change the expression of other markers. In contrast, inactivation of gdi-1 suppressed the expression of all three Rabs, and inactivation of M57.2, the enzyme that anchors Rab proteins to vesicles, caused diffuse GFP accumulation of all three Rabs, suggesting in both cases that all vesicular trafficking is disrupted. Both gene inactivations of the HOPs complex (B0303.9 and vps-16) caused luminal accumulation of late endosomes (RAB-7∷GFP) but not other Rabs, confirming a disruption of late endosomes (summarized in Supplementary Fig. S6). Two proteins known to be sorted through the endosome also change their localization. VIT-2∷GFP labels yolk protein (Supplementary Fig. S7), which is normally sorted into oocytes (Grant and Hirsh 1999), and a signal sequence GFP (ssGFP) (Supplementary Fig. S8), which is expressed in body wall muscles, secreted into the pseudocoelom, and finally endocytosed and degraded by coelomocyte scavenger cells (Supplementary Fig. S8A,B; Fares and Greenwald 2001). Five gene inactivations caused improper sorting of vitellogenin: gdi-1, cua-1, M57.2, C31H2.1, and F30A10.6 (Supplementary Fig. S7C–I; Supplementary Table S6), and most endocytosis-annotated gene inactivations suppressed coelomocyte vesicular fluorescence (Supplementary Table S6). Inactivation of vrk-1 was the only instance where an endocytosis-annotated gene did not alter any of the molecular markers for endocytosis. VRK-1 has weak similarity to the S. cerevisiae PKH2 (E−12), a serine/threonine protein kinase involved in a sphingolipid-mediated signaling pathway that controls endocytosis, consistent with the possibility that vrk-1 affects aging independent of vesicular trafficking. Inactivating daf-16 does not change VIT-2∷GFP or ssGFP localization or levels.
Discussion
Of the progeric gene inactivations identified in our screen, most enriched are genes annotated to mediate vesicle sorting. For example, compared with loss of daf-16, inactivation of Y65B4A.3 caused the greatest acceleration of daf-2 aging. Y65B4A.3 is homologous to human charged multivesicular body protein 6 and the myristolyated subunit of yeast ESCRT-III, the endosomal sorting complex required for transport of transmembrane proteins into the multivesicular body pathway to the lysosomal/vacuolar lumen (Babst et al. 2002), implying that inactivation of Y65B4A.3 inhibits normal vesicular trafficking to vacuoles/lysosomes. Sixteen other endocytosis/vesicular trafficking-related genes also shortened daf-2 life span and increased the rate of aging when inactivated, and nine of those strongly increased the rate of aging without increasing IMR: C31H2.1, T27F7.1, gdi-1, B0303.9, rab-7, CD4.4, Y47G6A.18, F30A10.6, and wwp-1.
A significant fraction of the genes that shorten daf-2 life span are annotated to mediate endocytotic trafficking to the lysosome (Fig. 4B–R, black circles). Ubiquitinated membrane proteins such as activated growth factor receptors or misfolded proteins are sorted through several vesicles, which involve multiple protein complexes prior to their ultimate degradation in the lysosome. Prior to degradation, plasma membrane proteins are ubiquitinylated by an E3 ubiquitin ligase. wwp-1 is the ortholog to Su(dx)/Itch, Nedd4 E3 ubiquitin ligases, which is required for LIN-12 degradation (Shaye and Greenwald 2005) and whose inactivation increased the rate of aging of daf-2 mutant animals. During vesicular trafficking from the plasma membrane to the lysosome, proteins are shuttled through early to late endosomes, where three ESCRT protein complexes function stepwise to sort ubiquinated membrane proteins into multivesicular bodies (MVB) for degradation. Significantly, five genes encoding subunits in the ESCRT complexes are required for daf-2 extension of life span and increased the rate of aging upon inactivation—CD4.4 (ESCRT-I), C27F2.5 (ESCRT-II), tag-318 (ESCRT-II), T27F7.1 (ESCRT- III), and Y65B4A.3 (ESCRT-III) (for review, see Slagsvold et al. 2006)—suggesting the ESCRT complexes are a major node whose disruption promotes accelerated aging. B0303.9 and vps-16 encode subunits of the HOPS complex, which mediates docking and fusing of MVBs with lysosomes. At each step, membrane vesicles are transported to the next compartment through various small GTPase Rab proteins. MVB trafficking from the late endosome to the lysosome is specifically guided by RAB-7, inactivation of which increases the rate of aging of daf-2 mutant animals. Two other proteins found in late endosomes, PTR-23 and CUA-1, also accelerate the aging of daf-2 mutant animals when inactivated—albeit the latter is slightly pleiotropic, with an IMR increased ∼30-fold. ptr-23 encodes a patched protein with high similarity to NPC1 (Niemann-Pick disease), and cua-1 encodes the ortholog of ATP7A/MNK with high similarity to ATP7B (Wilson disease). Three genes known to play a role in Rab function were necessary for daf-2 extended life span: M57.2, C31H2.1, and gdi-1. M57.2 is a geranylgeranyl transferase whose enzymatic action anchors Rab proteins. After vesicular transport from the donor to acceptor compartment, Rab proteins are recycled: First, Rab is inactivated and readied for recycling by a GTPase-activating protein (GAP), such as the probable Rab-GAP C31H2.1. Subsequently, a guanine nucleotide dissociation inhibitor (GDI), gdi-1, transports the inactive Rab from the acceptor membrane to the donor membrane. Y47G6A.18 is orthologous to the Golgi phosphoprotein 3. The Golgi complex sorts and modifies proteins exported from the endoplasmic reticulum. This protein is a peripheral membrane protein of the Golgi stack and may have a regulatory role in Golgi trafficking. F30A10.6 is orthologous to SAC1, a lipid phosphoinositide phosphatase of the ER and Golgi that is involved in protein trafficking and secretion (Whitters et al. 1993; Hughes et al. 2000; Schorr et al. 2001). Collectively, these results reveal a central role in vesicular trafficking to lysosomes and protein turnover in aging.
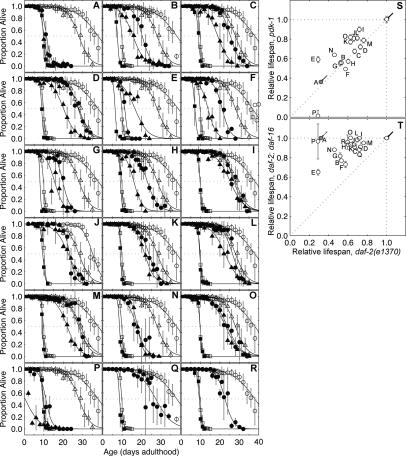
Figure 4.
Vesicular trafficking to the lysosome and protein sorting is required for decreased insulin/IGF1 signaling to extend life span. daf-2(e1370) (circles), pdk-1(sa680) (triangles), or daf-2(e1370);daf-16(mgDf47) (squares) animals on dsRNA to empty vector control (white symbols, A–R) or to daf-16 (A), CD4.4 (B), C27F2.5 (C), F17C11.8 (D), Y65B4A.3 (E), T27F7.1 (F), rab-7 (G), M57.2 (H), C31H2.1 (I), gdi-1 (J), vps-16 (K), B0303.9 (L), wwp-1 (M), vrk-1 (N), ptr-23 (O), cua-1 (P), Y47G6A.18 (Q), and F30A10.6 (R) (black symbols). (S) Relative life span (to empty vector control) of daf-2(e1370) (X-axis) versus pdk-1(sa680) (Y-axis) for each endocytosis gene inactivation. Empty vector control is indicated by a black arrow, daf-16 RNAi is indicated by a gray circle and gray arrow. (T) Relative life span (to empty vector control) of daf-2(e1370) (X-axis) versus daf-2(e1370);daf-16(mgDf47) (Y-axis) for each endocytosis gene inactivation. Empty vector control is indicated by a black arrow, daf-16 RNAi is indicated by a gray circle and gray arrow.
Examining the daf-2 pathway specificity by comparing the relative degree of life span shortening in daf-2 (Fig. 4A–R, black circles) to daf-2;daf-16 double-mutant animals (Fig. 4A–R, black squares) revealed that genes functioning in vesicular trafficking to lysosomes were specific to the daf-2 pathway, while the rest function in parallel to converge on the daf-2 pathway (Fig. 4T). The strict daf-2 pathway-specific members were vps-16, B0303.9, ptr-23, cua-1, F30A10.6, M57.2, and C32H2.1. Importantly, all of the remaining genes involved in endocytotic trafficking still shorten daf-2 life span to a greater degree than in daf-2; daf-16 and (with the exception of vrk-1) have no increase in IMR, implying function in a parallel pathway. In summary, this evidence demonstrates that aging-related genes involved in trafficking to lysosomes can be strictly placed in the daf-2 pathway when genes functioning closer to the lysosome are inactivated.
The shortened life span of these gene inactivations suggests that proper endocytosis is required for the extension of life span induced by decreased insulin/IGF1 signaling. One model for the role of endocytosis in aging is recycling of damaged cellular components. But the age pigment accumulation varies between these gene inactivations: Inactivation of some endocytosis genes causes accelerated accumulation of age pigment, whereas other inactivations caused less pigment accumulation. Thus, the accumulation of aging pigments during aging is a marker of the endocytotic decline, but some sorting defects also lead to less pigment accumulation. These data point to the endocytosis defects as key in aging, not age pigment accumulation per se.
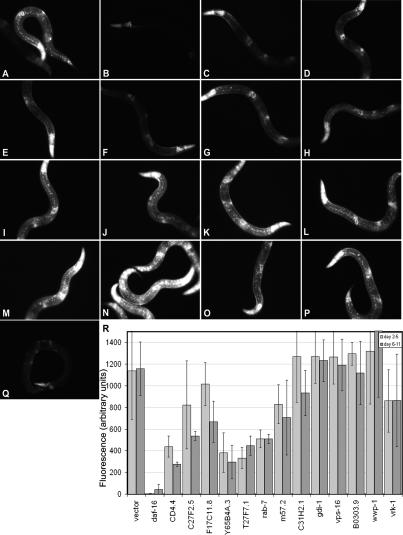
Many of the genes involved in vesicular trafficking to lysosomes that are necessary for decreased insulin/IGF1 signaling to extend C. elegans life span were also required for induction of sod-3. Specifically, all five subunits in the ESCRT complexes—CD4.4 (I), C27F2.5 (II), F17C11.8 (II), Y65B4A.3 (III), and T27F7.1 (III)—were required for elevation of sod-3 levels (Fig. 5C–G). Interestingly, the Rab effectors required for a daf-2 mutation to extend life span failed to affect sod-3 (Fig. 5I–K,R), as did other putative endocytosis genes (Fig. 5L–P,R). However, inactivation of M57.2, C31H2.1, or gdi-1 is not equivalent to inactivation of rab-7. Disruption of the former results in a loss of specificity in “all” vesicular trafficking, while inactivating the latter specifically disrupts vesicular trafficking to lysosomes. In this regard, loss of rab-7 is functionally most equivalent to disruption of the normal function of the ESCRT complex: In either case, membrane proteins targeted for lysosomal degradation would fail to be degraded. There are two general models for why defective endocytosis may shorten the life span of a daf-2 mutant. First, during aberrant vesicle sorting, signaling in the insulin/IGF1 pathway may no longer depend on engagement of the insulin/IGF1 receptor itself; for example, the PDK-1 or AKT-1/2 kinases may be released from their normal dependence on PIP3 signals. sod-3∷GFP expression is suppressed in many instances, suggesting that signaling downstream from the insulin/IGF1 receptor is activated in these animals. However, nearly all of the progeric endocytosis gene inactivations accelerate aging in pdk-1 mutant strains (Fig. 4B–R, black triangles; summarized in Fig. 4S), suggesting that it is not the missorting of PDK-1 signaling complexes that causes the progeria in a daf-2 mutant. These data favor a model of increased fidelity of essential endocytotic pathways extending longevity under conditions of low insulin/IGF1 signaling.
Figure 5.
Disruption of normal vesicular trafficking to the lysosome blocks sod-3 induction in daf-2 mutant animals. A representative daf- 2(e1370);sod-3∷GFP day 7 (adulthood) animal on RNAi to vector (A), daf-16 (B), CD4.4 (C), C27F2.5 (D), F17C11.8 (E), Y65B4A.3 (F), T27F7.1 (G), rab-7 (H), M57.2 (I), C31H2.1 (J), gdi-1 (K), vps-16 (L), B0303.9 (M), wwp-1 (N), vrk-1 (O), ptr-23 (P), and cua-1 (Q). (R) Quantification of average GFP body fluorescence between the nerve ring and vulva. (Light gray) Pooled animals scored between day 2 and 5 of adulthood; (dark gray) pooled animals scored between days 5 and 11.
Alternatively, vesicle sorting may be required for the downstream processes triggered by low insulin/IGF1 signaling; for example, autophagy of cellular components under low nutrient conditions. The autophagy genes bec-1, atg-7, and atg-12 are required for the extension of life span by daf-2(e1370) (Melendez et al. 2003; Hars et al. 2007). However, bec-1 scored as developmentally pleiotropic in our primary screen, and neither M7.5 (atg-7) nor B0336.8 (atg-12) were identified in our screen. Of the other genes implicated in autophagy, T22H9.2, Y60A3A.1 (unc-51), C32D5.9 (lgg-1), ZK593.6 (lgg-2), and R07G3.3 were tested and only F41E6.13 (atgr-18) slightly shortened daf-2 life span.
Several of the progeric genes identified in our screen have been identified in previous screens. hsf-1 is a major progeria gene found in our screen that converges with the daf-2 pathway. Inactivation of hsf-1 accelerates aging (Hsu et al. 2003; Morley and Morimoto 2004) to a similar extent as loss of daf-16. Loss of heat-shock factor (hsf-1), the primary transcription factor regulating stress-inducible gene expression and protein folding homeostasis, increased the rate of daf-2 aging more than fivefold, with a severe shortening of daf-2 life span (mean life span, 10.2 d). In addition to hsf-1 and pnk-1, we found two known aging genes that increased the rate of aging and were daf-2 pathway specific when inactivated: smk-1 (F41E6.3) and aak-2. SMK-1 is a component of insulin/IGF1 signaling that acts downstream from DAF-16 to regulate transcriptional specificity (Wolff et al. 2006). aak-2 encodes the AMPK α subunit and functions as a “fuel sensor” in the control of energy metabolism (Curtis et al. 2006).
While very few human progeriod syndromes have been assigned molecular causes, a mutation that disrupts a normal splice in human lamin A/C to induce inappropriate farnesylation causes Hutchison Gilford Syndrome, a devastating progeriod disease (Eriksson et al. 2003). Our screen also implicates nuclear lamin in aging. A C. elegans homolog of nuclear lamin, lmn-1 (DY3.2), emerged as a progeric gene inactivation that affects the life span of a daf-2 mutant more than wild type or a daf-2; daf-16 double mutant (Table 1). lmn-1 is approximately equally distant from human lamin B or human lamin A/C (probability of about E−51) and one of three C. elegans lamin genes that are equivalently homologous to a human lamin A/C query. None of the other paralogs of lmn-1 scored highly in our progeria screen. LMN-1 forms a nuclear ring in C. elegans that decays as animals age and decays more slowly in daf-2 mutant animals, and inactivation of lmn-1 in wild type shortens life span, all consistent with our observations (Haithcock et al. 2005).
Fat synthesis pathways also emerged as a daf-2 pathway-specific longevity axis. F28D1.9 is a very-long-chain acyl-CoA synthetase/fatty acid transporter and the only gene exclusively functioning within the daf-2 pathway with a mean life span and rate of aging similar to loss of daf-16. This suggests that metabolism of very-long-chain fatty acids is essential for the long life span of daf-2. Those metabolites could constitute hormonal signals to nuclear hormone receptors such as daf-12 ligands (Fisher and Lithgow 2006; Broue et al. 2007; Gerisch et al. 2007), or germline aging signals (Berman and Kenyon 2006), or a switch in metabolic pathway usage that in turn confers longevity. Consistent with a role for fatty acid metabolism in control of life span, RNAi of pnk-1 dramatically shortened daf-2 mutant adult life span (mean life span, 22.3 d) and increased the rate of aging almost threefold (2.88). pnk-1 (C10G11.5) is one of two C. elegans pantothenate kinases, the rate-limiting enzymes in coenzyme A synthesis, and is up-regulated in daf-2(m) animals to regulate fat storage (Lee et al. 2003). pnk-1 previously has been reported to mediate daf-2 mutant increased life span and is likely to be a direct DAF-16 target gene (Lee et al. 2003).
Several genes annotated to act in transcription or translation also increased the rate of daf-2 aging when inactivated. The C. elegans homolog of RAP74 is C01F1.1, the largest subunit of the transcription factor IIF (TFIIF), which is essential for accurate initiation of transcription and stimulates elongation by RNA polymerase II (Gaiser et al. 2000). Similarly, inactivation of either Y119C1B.8/tag-332 (subunit BDF1 of the TFIID complex) or Y56A3A.1/ntl-3 (ortholog of NOT3/5, a global transcriptional regulator with roles in transcription initiation and elongation and in mRNA degradation) accelerated aging. Additionally, inactivation of F55B12.4, a tRNA nucleotidyltransferase/poly(A) polymerase, also reduced the MRDT of daf-2(e1370) animals. Surprisingly, the relative IMR was similar to control animals, so that the median life span of animals after inactivating any of these genes was between 22 and 30 d—significantly shorter than the daf-2 life span of 36.9 d, suggesting that disrupting these processes accelerates aging rather than making animals sick.
Given that many microRNAs (miRNAs) are regulated late in adult life span, it may be significant that genes annotated to act in miRNA and small interfering RNA (siRNA) pathways are essential for long life span in both daf-2 and wild-type animals (Boehm and Slack 2005; Ibanez-Ventoso et al. 2006). This suggests that these small RNAs are important in the aging process. Comparing the 104 genes identified in this screen against three other RNAi screens—enhancement of the mg279 weak allele of let-7 to identify factors required for miRNA activity, and two independent screens to identify genes required for siRNA—showed a significant overlap with our screen (D. Parry, S. Kennedy, R. Kamath, H. Gabel, and J. Kim, pers. comm.). These annotations appeared at two to three times the frequency expected by chance.
Previous studies have focused on identifying gene inactivations that extend life span. Our screen used detailed demography, molecular markers, and a set of insulin/IGF1 signaling tester strains to filter progeric gene inactivations for specific acceleration of aging. Many of the genes identified in this study are necessary for the long life span of C. elegans insulin/IGF1 signaling-deficient mutants, and because they are conserved may act in homologous pathways in other animals. Sixty-three percent of the genes have orthologs in other organisms (BIOMART) (data not shown). The activities of these genes may also be regulated—for example, by insulin/IGF1 signaling—either transcriptionally by DAF-16 (and in other animals by the orthologous FOXO factors) or via phosphorylation of these protein; for example, by the PDK and AKT kinases, which are also highly conserved. Of the 104 genes identified, 4% have DAF-16-binding sites conserved to Caenorhabditis briggsae, suggesting that they constitute direct DAF-16 targets. These genes need not act proximally to DAF-16; our functional genomic screen identified indirect targets of insulin/IGF1 signaling as well as other aging pathways. For those genes that mediate long life span in parallel to the insulin/IGF1 signaling pathway, many (71%) are also conserved and may be regulated by converging or other pathways. The activities of both the insulin/IGF1 pathway genes and the parallel pathway genes may decline during aging so that the gene inactivations induced in this study could cause progeria by accelerating a normally time-regulated program.
Materials and methods
Strains
The following C. elegans strains were used in these studies: wild-type N2 Bristol, daf-2(e1370), daf-2(e1370); daf-16(mgDf47), daf- 2(e1370); muIs84 (sod-3∷GFP), pdk-1(sa680), unc-119(ed3);pwIs72 [vha6p∷GFP∷rab-5 + unc-119(+)], unc-119(ed3);pwIs170 [vha6p∷ GFP∷rab-7 + Cb unc-119(+)], unc-119(ed3);pwIs206 [vha6p∷ GFP∷rab-10 + Cb unc-119(+)], unc-119(ed3);pwIs503 [vha6p∷ mans∷GFP, Cb unc-119(+)], mgIs43 [ges-1p∷GFP], arIs37 [pmyo-3∷ssGFP], and sqt-1(sc103);bIs1 [vit-2∷GFP + rol-6(su1006)].
RNAi life span screen
The large-scale RNAi screen was carried out as described (Fraser et al. 2000) with slight modifications. Each RNAi colony was grown overnight in Luria broth with 50 μg mL−1 ampicillin and then seeded onto 24-well RNAi agar plates containing 5 mM isopropylthiogalactoside to induce dsRNA expression overnight at room temperature. About 25 synchronized daf-2(e1370) L1s were added to each well and allowed to develop to L4 at 15°C, then 5-fluorodeoxyuridine (FUDR) solution was added to the RNAi plates and the plates were moved to 25°C (day = 0). Survival was measured ∼22 and 25 d later, both by flooding the wells with water and gentle prodding with a platinum wire; animals that failed to move under either treatment were scored as dead. All of the RNAi clones identified in our screen were verified by DNA sequencing and subsequent BLAST analysis to confirm their identity. In eight instances out of hundreds identified correctly, the sequence result failed to match the expected gene identity and these RNAi clones were removed from further analysis. All annotations of RNAi clones/gene inactivations were according to WormBase.
Life span assay
Instead of following mortality in one population of animals over time, enough multiple replica sets were created so that one set could be scored, on average, every other day. Similar to the primary screen, each RNAi colony was grown overnight in Luria broth with 50 μg mL−1 ampicillin and then seeded onto 24-well RNAi agar plates containing 5 mM isopropylthiogalactoside to induce dsRNA expression overnight at room temperature. About 25 synchronized L1s were added to each well and allowed to develop to L4 at 15°C, then FUDR solution was added to the RNAi plates and the plates were moved to 25°C (day = 0). For each 24-well plate, one well contained bacteria expressing dsRNA to an empty vector and another to daf-16. At each time point, one replica in the set was measured by both flooding the wells with water and gentle prodding with a platinum wire; animals that failed to move under either treatment were scored as dead. At each subsequent time point, survival in a different replica set was measured. The survival proportion at each time point for every gene inactivation was obtained from three to five independent experiments.
Reproductive fitness
Each RNAi colony was grown overnight in Luria broth with 50 μg mL−1 ampicillin, concentrated 10×, and then seeded onto 6-cm agar plates containing 5 mM isopropylthiogalactoside to induce dsRNA expression overnight at room temperature. About 50 synchronized daf-2(e1370) L1s were added to each plate and allowed to develop at 15°C, and progeny were counted daily (eggs, L1, L2, or greater) until progeny reached the L4 stage.
Gompertz and logit fitting
The Gompertz model for age-specific mortality M = M0eGa, with initial mortality M0, aging coefficient G, and age a, can be shown to give the proportion dead P = 1 − exp[(M0/G)(1 − eGa)]. In the logit model for proportion dead, P = eb+ca/(1 + eb+ca), the parameters b and c are analogous to, but different than, log(M0) and G in the Gompertz model. Reflecting this limited analogy, we refer to b and c as initial mortality and the aging coefficient, respectively.
When analyzing data for a particular clone, sample proportions from different trials at the same age were pooled using population weighting. We performed population-weighted nonlinear least-squares Gompertz model fits using a Trust-Region Reflective Newton algorithm implemented by the Matlab fit() function (The MathWorks, Inc.). We performed population-weighted logit fits using the Matlab glmfit() function. We estimated the LD50, or age at which P = 0.5, given by log[1 + log(2)(G/M0)]/G in the Gompertz model or −b/a in the logit model. When neither Gompertz nor logit models could be fit (most commonly due to lack of data in the transition region due to rapid changes in proportion dead relative to scoring frequency), median life span was determined from a linear fit to proportions 0 < P <1.
MRDT analysis
We tested whether the MRDT = log(2)/G differed significantly for each clone (C) from the vector control (V). We performed t-tests using t = (GC − GV)/[(sG,C2/DOFC) + (sG,V2/DOFV)], where sG2 is the estimated variance of G, and DOF is the remaining degrees of freedom after fitting a Gompertz model (number of proportions − 2). P-values were estimated using overall degrees of freedom DOF = DOFC + DOFV − 2, corrected using the Bonferroni step-down method, and considered significant for P < 0.05. We also computed the relative rate of aging (MRDTV/MRDTC = GC/GV), using Monte-Carlo techniques (N = 1000 random samples) to estimate its variability from the estimated uncertainty of GC and GV.
Activity analysis
The Gompertz model, but not the logit model, constrains the sample proportion to be zero at time 0 (start of adulthood); this boundary condition, while acceptable for mortality data (proportion dead), may be inconsistent with movement data (proportion inactive), such as in the case of sickness. We analyzed paired movement and mortality data using the logit model. We estimated the active proportion of adult life span in two ways: first as the ratio of movement LD50 to mortality LD50, RL, and second as the ratio of integrals (movement/mortality) RI, a more accurate approach for animals with a short active span. For each proportion fit π(a), we used an adaptive Simpson quadrature method [Matlab quad() function] to compute the integral S = ∫ [1 − π(a)]da over [0,A], with A an arbitrary value greater than the maximum life span. Here, S represents the expected life span for mortality data, or the expected active period for movement data.
Monte-Carlo techniques were used to estimate the variability of R based on the estimated uncertainties in the LD50 values for RL, and in the S values for RI. In the latter case, for each set of clone data, we generated 1000 π(a) curves based on the estimated uncertainties of parameters b and c, and calculated S for each π(a), allowing the variability in R to be estimated from the distributions of the S values.
Using a similar approach as the MRDT analysis, we performed t-tests on paired movement and mortality LD50 and S values. R ratios for a given clone were considered significantly <1 if P-values, corrected using the Bonferroni method, were significant (<0.05) for both LD50 and S t-statistics. This procedure eliminated clones with large uncertainties in LD50 and S values, a subset of which might conceivably have the physically impossible R > 1 (active period longer than life span) due to noisy data, small-N statistics, and incongruence between data and the logit model. In cases of integral nonconvergence or large activity ratio uncertainties failing the aforementioned test, we estimated the activity ratio, with unquantified uncertainty, as the quotient of median active span to median life span.
Pathway specificity analysis
We defined daf-16 pathway independence as equivalent relative reduction in life span for a given gene inactivation in both daf-2(e1370) and daf-2(e1370);daf-16(mgDf47) mutants. On a plot with single- and double-mutant relative life spans as abscissa (X) and ordinate (Y), Y = X represents independence, Y = 1 represents daf-16 pathway specific, and RNAi vector controls lie at the point (1,1). Gene inactivations were classified as specific for angles, relative to the control point, within 10° of the Y = X line, independent for angles within 10° of Y = 1, and parallel/convergent for intermediate values.
Accumulation of age pigment
To access whether a gene inactivation accelerated the accumulation of autofluorescent age pigment, each RNAi colony was grown overnight in Luria broth with 50 μg mL−1 ampicillin and then seeded onto six-well RNAi agar plates containing 5 mM isopropylthiogalactoside to induce dsRNA expression overnight at room temperature. Between 50 and 75 synchronized daf-2(e1370) L1s were added to each well and allowed to develop to L4 at 15°C, then FUDR solution was added to the RNAi plates and the plates were moved to 25°C (day = 0). Animals were followed longitudinally to day 15 of adulthood or later while feeding on bacteria corresponding to the RNAi clones that shorten life span, and populations were scored blindly both for relative increase in the intensity and spreading of age-associated autofluorescence within individual animals and within the whole population. Gene inactivations were classified as “strong” accelerated accumulation of age pigment if 100% of animals consistently scored positive from day 2 to 12 of adulthood. Similarly, gene inactivations with “moderate” premature age pigment accumulation had observable autofluorescence in >50% of animals from day 5 to 15 of adulthood. Moderate classification differed from strong based chiefly in the proportion of animals consistently scored as displaying accelerated age pigment accumulation and, to a lesser extent, intensity of autofluorescence. Gene inactivations resulting either in only a modest increase in autofluorescence over control, intense autofluorescence in <50% of the population, or accelerated age pigment after day 15 of adulthood were classified as weak (summarized in Tables 1–3). Age pigment was observed with a Stemi SV11 Zeiss fluorescent or a Discovery V12 Zeiss stereo microscope. To compare age pigment accumulation in wild-type (N2) versus daf-2 mutant animals (e1370 or m41) exposed to either daf-16 RNAi or empty vector control (as above), ∼50 animals under each condition were scored blindly on day 6 of adulthood at 25°C on the basis of whether they displayed autofluorescence under a dissecting GFP microscope.
Sod-3 expression
daf-2 mutant animals coexpressing a sod-3∷GFP reporter were fed dsRNA-expressing bacteria for each gene necessary for daf-2 life span extension, and animals were followed longitudinally (blind) from early adulthood to identify genes whose function are required for decreased insulin signaling to induce sod-3. Images of representative animals were taken at 37× magnification on a Discovery V12 Zeiss stereo microscope. Quantification of sod-3 expression levels was obtained using Axiovision release version 4.5 software (Zeiss) of the GFP signal between the pharynx and the vulva opening. Nonworm background fluorescence was subtracted, and the average fluorescence for both early and late adulthood was calculated (day 2–5 and 6–11, respectively). All adult animals were maintained at 25°C.
Acknowledgments
We thank the Ellison Medical foundation for funding the Marine Biology Laboratory, Biology of Aging course where the age pigment screen was conducted, and students of A.V.S.: Andrej Rotter, Adrienne Frostholm, Lenka Rossmeislova, Marsha Penner, Qunxing Ding, and Vidya Nukala. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. Additionally, we thank Emily Troemel, Alexander Soukas, Denise Chun, Barth Grant, and Boudewijn Burgering for providing some of the strains used in this study. We thank Alexander Soukas for critical reading of the manuscript, members of the Ruvkun laboratories for scientific discussions and support, and Andrea Carrano for scientific discussions. We are thankful to Devin Parry, Harrison Gabel, John Kim, Ravi Kamath, and Scott Kennedy for communication of their RNAi screens. This research was funded by NIH grant #5_R01_AG16636. A.V.S. would like to dedicate this work to his father, Dr. H. Vaughn Samuelson, who is retiring at the end of the year from DuPont after 42 years of continuous service in research and development.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1588907
References
- Apfeld J., Kenyon C., Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D., Estepa-Sabal E.J., Meerloo T., Emr S.D., Meerloo T., Emr S.D., Emr S.D. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A., Viswanathan M., Horvitz H.R., Guarente L., Viswanathan M., Horvitz H.R., Guarente L., Horvitz H.R., Guarente L., Guarente L. C. elegans SIR-2.1 interacts with 14–3–3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman J.R., Kenyon C., Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Bluher M., Kahn B.B., Kahn C.R., Kahn B.B., Kahn C.R., Kahn C.R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Boehm M., Slack F., Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Broue F., Liere P., Kenyon C., Baulieu E.E., Liere P., Kenyon C., Baulieu E.E., Kenyon C., Baulieu E.E., Baulieu E.E. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell. 2007;6:87–94. doi: 10.1111/j.1474-9726.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Brunk U.T., Terman A., Terman A. Lipofuscin: Mechanisms of age-related accumulation and influence on cell function. Free Radic. Biol. Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- Curtis R., O’Connor G., DiStefano P.S., O’Connor G., DiStefano P.S., DiStefano P.S. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Dillin A., Crawford D.K., Kenyon C., Crawford D.K., Kenyon C., Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., Robbins C.M., Moses T.Y., Berglund P., Moses T.Y., Berglund P., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: Coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C.E., Ruvkun G., Ruvkun G. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Fisher A.L., Lithgow G.J., Lithgow G.J. The nuclear hormone receptor DAF-12 has opposing effects on Caenorhabditis elegans lifespan and regulates genes repressed in multiple long-lived worms. Aging Cell. 2006;5:127–138. doi: 10.1111/j.1474-9726.2006.00203.x. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Martinez-Campos M., Sohrmann M., Ahringer J., Sohrmann M., Ahringer J., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Furuyama T., Nakazawa T., Nakano I., Mori N., Nakazawa T., Nakano I., Mori N., Nakano I., Mori N., Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser F., Tan S., Richmond T.J., Tan S., Richmond T.J., Richmond T.J. Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 Å resolution. J. Mol. Biol. 2000;302:1119–1127. doi: 10.1006/jmbi.2000.4110. [DOI] [PubMed] [Google Scholar]
- Garigan D., Hsu A.L., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Hsu A.L., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Fraser A.G., Kamath R.S., Ahringer J., Kenyon C., Kamath R.S., Ahringer J., Kenyon C., Ahringer J., Kenyon C., Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo R.S. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 2002;13:156–162. doi: 10.1016/s1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- Gems D., Riddle D.L., Riddle D.L. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B., Rottiers V., Li D., Motola D.L., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A., Rottiers V., Li D., Motola D.L., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A., Li D., Motola D.L., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A., Motola D.L., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A., Lehrach H., Mangelsdorf D.J., Antebi A., Mangelsdorf D.J., Antebi A., Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc. Natl. Acad. Sci. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstbrein B., Stamatas G., Kollias N., Driscoll M., Stamatas G., Kollias N., Driscoll M., Kollias N., Driscoll M., Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Golden T.R., Melov S., Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. 2004;3:111–124. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- Grant B., Hirsh D., Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock E., Dayani Y., Neufeld E., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J., Dayani Y., Neufeld E., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J., Neufeld E., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J., Feinstein N., Mattout A., Gruenbaum Y., Liu J., Mattout A., Gruenbaum Y., Liu J., Gruenbaum Y., Liu J., Liu J. Age- related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hars E.S., Qi H., Ryazanov A.G., Jin S., Cai L., Hu C., Liu L.F., Qi H., Ryazanov A.G., Jin S., Cai L., Hu C., Liu L.F., Ryazanov A.G., Jin S., Cai L., Hu C., Liu L.F., Jin S., Cai L., Hu C., Liu L.F., Cai L., Hu C., Liu L.F., Hu C., Liu L.F., Liu L.F. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Geloen A., Even P.C., Cervera P., Le Bouc Y., Dupont J., Ducos B., Leneuve P., Geloen A., Even P.C., Cervera P., Le Bouc Y., Ducos B., Leneuve P., Geloen A., Even P.C., Cervera P., Le Bouc Y., Leneuve P., Geloen A., Even P.C., Cervera P., Le Bouc Y., Geloen A., Even P.C., Cervera P., Le Bouc Y., Even P.C., Cervera P., Le Bouc Y., Cervera P., Le Bouc Y., Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Honda Y., Honda S., Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hsu A.L., Murphy C.T., Kenyon C., Murphy C.T., Kenyon C., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Huang C., Xiong C., Kornfeld K., Xiong C., Kornfeld K., Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Woscholski R., Cooke F.T., Patrick R.S., Dove S.K., McDonald N.Q., Parker P.J., Woscholski R., Cooke F.T., Patrick R.S., Dove S.K., McDonald N.Q., Parker P.J., Cooke F.T., Patrick R.S., Dove S.K., McDonald N.Q., Parker P.J., Patrick R.S., Dove S.K., McDonald N.Q., Parker P.J., Dove S.K., McDonald N.Q., Parker P.J., McDonald N.Q., Parker P.J., Parker P.J. SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 2000;275:801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C., Yang M., Guo S., Robins H., Padgett R.W., Driscoll M., Yang M., Guo S., Robins H., Padgett R.W., Driscoll M., Guo S., Robins H., Padgett R.W., Driscoll M., Robins H., Padgett R.W., Driscoll M., Padgett R.W., Driscoll M., Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Johnson T.E. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science. 1990;249:908–912. doi: 10.1126/science.2392681. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., Chang J., Gensch E., Rudner A., Tabtiang R., Gensch E., Rudner A., Tabtiang R., Rudner A., Tabtiang R., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum H.A., Liu Y., Ruvkun G., Tissenbaum H.A., Liu Y., Ruvkun G., Liu Y., Ruvkun G., Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Klass M.R. Aging in the nematode Caenorhabditis elegans: Major biological and environmental factors influencing life span. Mech. Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Larsen P.L. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1993;90:8905– 8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.Y., Hench J., Ruvkun G., Hench J., Ruvkun G., Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Lee S.S., Kennedy S., Tolonen A.C., Ruvkun G., Kennedy S., Tolonen A.C., Ruvkun G., Tolonen A.C., Ruvkun G., Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Li W., Boswell R., Wood W.B., Boswell R., Wood W.B., Wood W.B. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 2000;218:172–182. doi: 10.1006/dbio.1999.9593. [DOI] [PubMed] [Google Scholar]
- Li J., Tewari M., Vidal M., Lee S.S., Tewari M., Vidal M., Lee S.S., Vidal M., Lee S.S., Lee S.S. The 14–3–3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev. Biol. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J.R., Kenyon C., Berman J.R., Kenyon C., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K., Dorman J.B., Rodan A., Kenyon C., Dorman J.B., Rodan A., Kenyon C., Rodan A., Kenyon C., Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- McElwee J., Bubb K., Thomas J.H., Bubb K., Thomas J.H., Thomas J.H. Transcriptional outputs of the Caenorhabditis elegansforkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Seaman M., Eskelinen E.L., Hall D.H., Levine B., Eskelinen E.L., Hall D.H., Levine B., Hall D.H., Levine B., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Morley J.F., Morimoto R.I., Morimoto R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Kamath R.S., Ahringer J., Li H., Kenyon C., Ahringer J., Li H., Kenyon C., Li H., Kenyon C., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nakae J., Biggs W.H., Kitamura T., Cavenee W.K., Wright C.V., Arden K.C., Accili D., Biggs W.H., Kitamura T., Cavenee W.K., Wright C.V., Arden K.C., Accili D., Kitamura T., Cavenee W.K., Wright C.V., Arden K.C., Accili D., Cavenee W.K., Wright C.V., Arden K.C., Accili D., Wright C.V., Arden K.C., Accili D., Arden K.C., Accili D., Accili D. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G., Lee L., Tissenbaum H.A., Ruvkun G., Tissenbaum H.A., Ruvkun G., Ruvkun G. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Papini E., Satin B., Bucci C., de Bernard M., Telford J.L., Manetti R., Rappuoli R., Zerial M., Montecucco C., Satin B., Bucci C., de Bernard M., Telford J.L., Manetti R., Rappuoli R., Zerial M., Montecucco C., Bucci C., de Bernard M., Telford J.L., Manetti R., Rappuoli R., Zerial M., Montecucco C., de Bernard M., Telford J.L., Manetti R., Rappuoli R., Zerial M., Montecucco C., Telford J.L., Manetti R., Rappuoli R., Zerial M., Montecucco C., Manetti R., Rappuoli R., Zerial M., Montecucco C., Rappuoli R., Zerial M., Montecucco C., Zerial M., Montecucco C., Montecucco C. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J. 1997;16:15–24. doi: 10.1093/emboj/16.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S.D., Khazaeli A.A., Curtsinger J.W., Khazaeli A.A., Curtsinger J.W., Curtsinger J.W. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J. Gerontol. A Biol. Sci. Med. Sci. 2000;55:B381–B389. doi: 10.1093/gerona/55.8.b381. [DOI] [PubMed] [Google Scholar]
- Russell R.L., Seppa R.I., Seppa R.I. Genetic and environmental manipulation of aging in Caenorhabditis elegans. Basic Life Sci. 1987;42:35–48. doi: 10.1007/978-1-4613-1939-9_3. [DOI] [PubMed] [Google Scholar]
- Schorr M., Then A., Tahirovic S., Hug N., Mayinger P., Then A., Tahirovic S., Hug N., Mayinger P., Tahirovic S., Hug N., Mayinger P., Hug N., Mayinger P., Mayinger P. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 2001;11:1421–1426. doi: 10.1016/s0960-9822(01)00449-3. [DOI] [PubMed] [Google Scholar]
- Shaye D.D., Greenwald I., Greenwald I. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development. 2005;132:5081–5092. doi: 10.1242/dev.02076. [DOI] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H., Pattni K., Malerod L., Stenmark H., Malerod L., Stenmark H., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Terman A., Brunk U.T., Brunk U.T. Oxidative stress, accumulation of biological ‘garbage,’ and aging. Antioxid. Redox Signal. 2006;8:197–204. doi: 10.1089/ars.2006.8.197. [DOI] [PubMed] [Google Scholar]
- Terman A., Gustafsson B., Brunk U.T., Gustafsson B., Brunk U.T., Brunk U.T. Mitochondrial damage and intralysosomal degradation in cellular aging. Mol. Aspects Med. 2006;27:471–482. doi: 10.1016/j.mam.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Tu M.P., Epstein D., Tatar M., Epstein D., Tatar M., Tatar M. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell. 2002;1:75–80. doi: 10.1046/j.1474-9728.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J.R., De Vreese A., De Vreese A. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. FASEB J. 1995;9:1355–1361. doi: 10.1096/fasebj.9.13.7557026. [DOI] [PubMed] [Google Scholar]
- Vitelli R., Santillo M., Lattero D., Chiariello M., Bifulco M., Bruni C.B., Bucci C., Santillo M., Lattero D., Chiariello M., Bifulco M., Bruni C.B., Bucci C., Lattero D., Chiariello M., Bifulco M., Bruni C.B., Bucci C., Chiariello M., Bifulco M., Bruni C.B., Bucci C., Bifulco M., Bruni C.B., Bucci C., Bruni C.B., Bucci C., Bucci C. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 1997;272:4391–4397. doi: 10.1074/jbc.272.7.4391. [DOI] [PubMed] [Google Scholar]
- Wadsworth W.G., Riddle D.L., Riddle D.L. Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev. Biol. 1989;132:167–173. doi: 10.1016/0012-1606(89)90214-5. [DOI] [PubMed] [Google Scholar]
- Wang Y., Oh S.W., Deplancke B., Luo J., Walhout A.J., Tissenbaum H.A., Oh S.W., Deplancke B., Luo J., Walhout A.J., Tissenbaum H.A., Deplancke B., Luo J., Walhout A.J., Tissenbaum H.A., Luo J., Walhout A.J., Tissenbaum H.A., Walhout A.J., Tissenbaum H.A., Tissenbaum H.A. C. elegans 14–3–3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Whitters E.A., Cleves A.E., McGee T.P., Skinner H.B., Bankaitis V.A., Cleves A.E., McGee T.P., Skinner H.B., Bankaitis V.A., McGee T.P., Skinner H.B., Bankaitis V.A., Skinner H.B., Bankaitis V.A., Bankaitis V.A. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J. Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S., Ma H., Burch D., Maciel G.A., Hunter T., Dillin A., Ma H., Burch D., Maciel G.A., Hunter T., Dillin A., Burch D., Maciel G.A., Hunter T., Dillin A., Maciel G.A., Hunter T., Dillin A., Hunter T., Dillin A., Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., Kimura K.D., Lee M.S., Ruvkun G., Kimura K.D., Lee M.S., Ruvkun G., Lee M.S., Ruvkun G., Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]