Abstract
Thioredoxin (TRX) is a pleiotropic cellular factor that has thiol-mediated redox activity and is important in regulation of cellular processes, including proliferation, apoptosis, and gene expression. The activity of several transcription factors is posttranslationally altered by redox modification(s) of specific cysteine residue(s). One such factor is nuclear factor (NF)-κB, whose DNA-binding activity is markedly augmented by TRX treatment in vitro. Similarly, the DNA-binding activity of activator protein 1 (AP-1) is modified by a DNA repair enzyme, redox factor 1 (Ref-1), which is identical to a DNA repair enzyme, AP endonuclease. Ref-1 activity is in turn modulated by various redox-active compounds, including TRX. We here report the molecular cascade of redox regulation of AP-1 mediated by TRX and Ref-1. Phorbol 12-myristate 13 acetate efficiently translocated TRX into the HeLa cell nucleus where Ref-1 preexists. This process seems to be essential for AP-1 activation by redox modification because co-overexpression of TRX and Ref-1 in COS-7 cells potentiated AP-1 activity only after TRX was transported into the nucleus by phorbol 12-myristate 13 acetate treatment. To prove the direct active site-mediated association between TRX and Ref-1, we generated a series of substitution-mutant cysteine residues of TRX. In both an in vitro diamide-induced cross-linking study and an in vivo mammalian two-hybrid assay we proved that TRX can associate directly with Ref-1 in the nucleus; also, we demonstrated the requirement of cysteine residues in the TRX catalytic center for the potentiation of AP-1 activity. This report presents an example of a cascade in cellular redox regulation.
Keywords: redox regulation, disulfide cross-linking, nuclear translocation, mammalian two-hybrid assay
Increasing evidence has indicated that cellular redox status modulates various aspects of cellular events, including proliferation and apoptosis (1). TRX is a small, ubiquitous protein with two redox-active half-cystine residues, -Cys-Gly-Pro-Cys-, in an active center (2–5). TRX also is known as adult T-cell leukemia-derived factor that is involved in human T lymphotropic virus type I leukemogenesis (6, 7). TRX exists either in a reduced or oxidized form and participates in redox reactions through the reversible oxidation of this active center dithiol.
TRX serves functions inside (8–10) and outside (11–13) the cell. One of its intracellular functions is the facilitation of protein–nucleic acid interactions (3). In vitro and in vivo experiments showed that TRX augmented the DNA-binding and transcriptional activities of the p50 subunit of nuclear factor (NF)-κB by reducing Cys-62 in its DNA-binding loop (8, 9). Recently, direct physical association of an active-site mutant of TRX and an oligopeptide from NF-κB p50 was demonstrated by a nuclear magnetic resonance (NMR) study in vitro (14); in vivo direct association of TRX–NF-κB remains to be demonstrated. Redox regulation of Jun and Fos molecules has also been implicated. Various antioxidants strongly activate the DNA-binding and transactivation abilities of activator protein 1 (AP-1) complex (1, 15, 16). Interestingly, TRX enhances the DNA-binding activity of Jun and Fos, and this process requires other molecules such as a novel protein called redox factor 1 (Ref-1) (17, 18). Ref-1 was identified in a cell-free system as one of the factors restoring AP-1-DNA binding and was found to be identical to an AP endonuclease (17, 19). There has been no report describing whether TRX and Ref-1 directly associate or require any intervening factor(s) for their interaction. In this paper we studied the association of TRX and Ref-1 through the use of an in vitro cross-linking and a mammalian two-hybrid system, with various mutants of TRX, to investigate the interaction of TRX on AP-1-mediated transcription.
MATERIALS AND METHODS
Cells and Cell Culture.
HeLa and COS-7 cells were cultured in DMEM (GIBCO/BRL) supplemented with 10% fetal calf serum and antibiotics at 95% humidity/5% CO2 in air at 37°C.
Reagents.
Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma. Anti-human TRX mAb, 11-mAb, whose epitope has not been determined (10), was produced and provided by Fujirebio (Tokyo). Anti-human Ref-1 rabbit polyclonal antibody (C-20), which recognizes the epitope corresponding to amino acids 299–318, and anti-c-Fos mAb (6–2H-2F), which recognizes the epitope corresponding to the leucine zipper of c-Fos, were purchased from Santa Cruz Biotechnology.
Mutagenesis of TRX and Transfection.
Mutagenesis of human TRX was performed by a PCR-based technique (20), and authenticity of the sequences was verified by DNA sequencing (Fig. 1). The expression plasmids were introduced into the cells by LipofectAMINE reagent (GIBCO/BRL) according to the manufacturer’s instructions, using Opti-MEM (GIBCO/BRL).
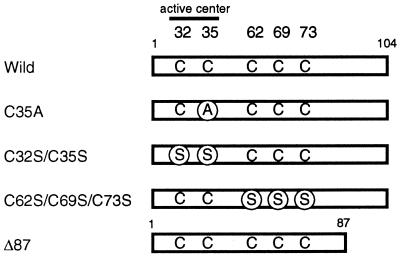
Figure 1.
Schematic representation of mutant constructs of TRX used in this experiment.
Luciferase Assay Using -73/+63 Col (Human Collagenase I)-LUC.
The −73/+63 Col-LUC was constructed with pGL2-Basic (Promega) and a fragment from −73/+63 Col in pBLCAT3 (21). HeLa cells were plated in 6-well plates at a density of 4 × 105 cells per well. The expression plasmids were introduced into the cells by use of LipofectAMINE reagent. In each transfection, 1 μg of the pCDSRα-TRXwild or -TRXC32S/C35S expression plasmid (6) and/or 1 μg of the pRc/CMV-Ref-1 expression vector with 1 μg of −73/+63 Col-LUC were used. The total amount of DNA was adjusted to 3 μg with pRc/CMV (Invitrogen). After incubation of 16 hr, PMA (30 ng·ml−1) was added and the cells were kept for 24 hr. The cells were harvested after 24 hr and luciferase activity was measured by use of a commercial assay system (Promega) with a luminometer. The relative fold induction of luciferase activity was calculated.
Expression of Recombinant Proteins in Bacteria.
Various TRX recombinant proteins were expressed in Escherichia coli as the N-terminal fused form with hexahistidine (6xHis) tag using the pQE30 expression plasmids (Qiagen, Chatsworth, CA). E. coli strain M15 [pRep4] was transformed with each pQE–TRX expression vector. E. coli were grown at 37°C with vigorous shaking and were treated for 3 hr with 1 mM isopropyl β-d-thiogalactoside, and the pelleted cells were lysed in PBS containing 1 mg·ml−1 lysozyme, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 50 μM 7-amino-1-chloro-3-tosylamido-2-heptanone, and 0.8 mM imidazol and sonicated, and was clarified by centrifugation. The supernatant containing 6xHis–TRX was loaded onto a Ni2+-NTA (nitrilo-tri-acetic acid) agarose column. The column was washed, and the fusion protein was eluted by PBS containing 80 mM imidazol. The eluted protein was dialyzed against PBS (pH 7.3) containing 2 mM DTT. Human Ref-1 was expressed based on the pDS56ref-1 (a gift form S. Xanthoudakis) (18) as a 6xHis-tagged protein by use of the same protocol as that used for the expression of TRX, except that the Ref-1 eluate was dialyzed against a storage buffer [50 mM sodium phosphate, pH 7.3/50 mM NaCl/5 mM MgCl2/1 mM EDTA, 5% (vol/vol) glycerol] containing 1 mM DTT. Truncated rat 6xHis-tagged recombinant c-Fos proteins, wbFos and its cysteine-to-serine mutant, wbFos (C1-S), were expressed and purified as described, based on pDS56wbFos and pDS56wbFos (gifts form T. Curran) (C1-S) (18).
In Vitro Disulfide Cross-linking Interaction Assay.
In the cross-linking assay, 6xHis–TRXs (200 ng), 6xHis–Ref-1 (200 ng) and 6xHis–Fos (200 ng) were incubated for 30 min at room temperature with 1 mM DTT. Then they were mixed and incubated in the presence of 10 mM diamide for 30 min at room temperature in 20 μl of PBS (pH 7.3). The reaction mixture was denatured for 5 min at 90°C in dissociation buffer with or without 2-mercaptoethanol (1%). Each mixture was applied to a 15% SDS/PAGE and electrophoresed (18). After electroblotting, polyvinylidene difluoride membranes (Millipore) were blocked, then incubated with primary antibodies followed by incubation with horseradish peroxidase-conjugated anti-IgG antibody (Amersham). The antigens on the membrane were visualized with the ECL Western blot detection kit (Amersham).
Indirect Immunofluorescence Cell Staining.
Cells were fixed with 3.7% paraformaldehyde in PBS containing 10% fetal calf serum for 20 min at room temperature followed by permeabilization for 10 min using 0.2% (wt/vol) Triton X-100 in PBS. After incubation (1 hr) with primary antibodies (for TRX, monoclonal 11-mAb; for Ref-1, polyclonal antibody, C-20) slides were incubated (1 hr) with either dye-labeled anti-mouse or anti-rabbit IgG. Slides with stained cells were mounted in 90% glycerol with 1 mg·ml−1 p-phenylenediamine. Cells were examined using a confocal microscopy, MRC 600 (Bio-Rad).
Mammalian Two-Hybrid Assay (22).
A complementary DNA of TRX, its mutants, or Ref-1 were fused in frame to pCMX-GAL4 (23) or pCMX-VP16 (24) (gifts from K. Umesono, Nara Institute of Science and Technology). The COS-7 cells were plated in 6-well plates at a density of 2 × 105 cells per well. The expression plasmids were introduced into the cells by LipofectAMINE reagent. In each transfection, 1 μg of GAL4-fused plasmid and 1 μg of viral protein 16 (VP16)-fused plasmid were used together with 1 μg of reporter construct, ptk-GALpx3-LUC (a gift from K. Umesono, Nara Institute of Science and Technology) made from pUC8 by inserting three copies of Gal4-binding site and a minimal thymidine-kinase promoter-luciferase expression cassette (this reporter plasmid has no AP-1-binding site which may influence the expression of luciferase). As an internal control, 1 μg of β-galactosidase expression plasmid was used. The cells were harvested after 30 hr, and luciferase activity was determined. The relative fold induction of luciferase activity was calculated by normalizing to the β-galactosidase activity.
RESULTS
Transient Expression of TRX and/or Ref-1 Activate AP-1 in PMA-Stimulated HeLa Cells.
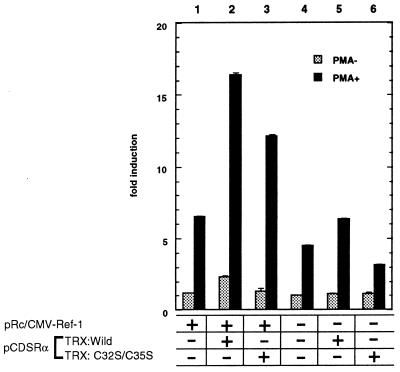
We examined whether overexpressed TRX and/or Ref-1 resulted in transcriptional activation of AP-1 in vivo. Thus we assayed the activity of the human collagenase I promoter, which contains an AP-1-binding site, by determining the luciferase activity in HeLa cells (Fig. 2). Transient overexpression of TRX and/or Ref-1 had only a marginal effect for induction of luciferase activity. However when treated by PMA, the luciferase activity increased about 4-fold (lane 2) over the mock transfection (lane 4). Expression of either wild-type TRX (lane 5) or Ref-1 (lane 1) alone resulted in less than 1.5-fold or 2-fold activation, respectively. Expression of TRXC32S/C35S, which lacked reducing activity, caused only marginal induction (lane 6). Thus, overexpressed TRX and Ref-1 seemed to potentiate the AP-1 transcriptional activity.
Figure 2.
Effect of transient expression of TRX and/or Ref-1 on AP-1 transactivation in HeLa cells with or without PMA treatment. In each transfection, 1 μg of the pCDSRα-TRXwild (lanes 2 and 5) or -TRXC32S/C35S (lanes 3 and 6) expression plasmid and/or 1 μg of the pRc/CMV–Ref-1 (lanes 1–3) expression vector with 1 μg of −73/+63 Col-LUC were used. The total amount of DNA was adjusted to 3 μg with pRc/CMV (Invitrogen). After incubation for 16 hr, PMA (30 ng·ml−1) was added and the cells were kept for 24 hr. The cells were harvested after 40 hr, and luciferase activity was determined. The results are the means ± SE of three experiments (each in duplicate) and presented as fold increases in luciferase activity over the baseline seen in lane 4 without PMA treatment.
PMA Treatment Translocated TRX from the Cytoplasm into the Nucleus in HeLa Cells.
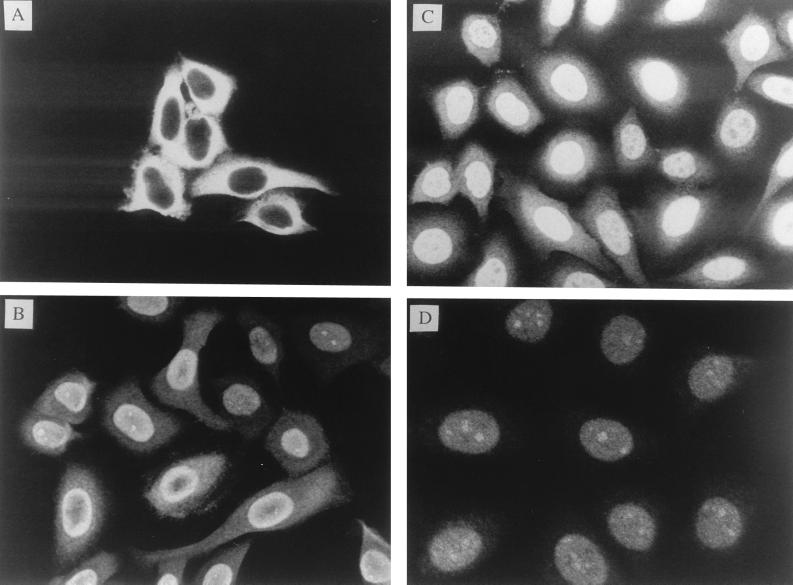
We next examined the intracellular locations of TRX and Ref-1. We analyzed HeLa cells without or with PMA treatment (to induce transcriptional activation of AP-1) (15, 21), by an indirect immunofluorescence method, using antibodies raised against TRX or Ref-1. As reported elsewhere, Ref-1 was localized in the nucleus and remained there after PMA stimulation (Fig. 3D) (17, 25). In contrast, TRX was located predominantly in the cytoplasm before PMA stimulation (Fig. 3A) but was translocated into the nucleus after 1 hr of PMA treatment (Fig. 3B); this process was almost complete in 24 hr (Fig. 3C).
Figure 3.
Effect of PMA treatment on subcellular distribution of TRX and Ref-1 in HeLa cells. Indirect immunofluorescence labelling, using anti-TRX antibody (A–C) or anti-Ref-1 antibody (D). (A) Before PMA treatment. (B) After 1-hr treatment. (C and D) After 24-hr treatment.
TRX Associates with Ref-1 through Cysteines of Catalytic Center in Vitro.
As suggested by Holmgren (3), an oxido-reductase like TRX should form an intermediate through disulfide linkage. Therefore, we reasoned that it would be possible to trap the transient physical association using cross-linking reagent(s) such as diamide, which converts free sulfhydryls to disulfides by cysteine oxidation (26, 27). To determine which of the five cysteines in TRX is responsible for intermolecular disulfide bond formation, three TRX mutants, which contained cysteines-to-serines or cysteine-to-alanine substitution(s), were purified as the 6xHis-tagged form. A series of experiments was performed with the 6xHis-tagged Ref-1 and the various mutants of TRX (Fig. 1). It is known that TRXC62S/C69S/C73S retains its reducing activity (28, 29), while substitution mutants (TRXC35A, TRXC32S/C35S) at either Cys-32 or Cys-35 lose their in vitro reducing activity (3). In spite of the loss of reducing activity, TRXC35A is able to involve the covalent mixed disulfide intermediate formation between its Cys-32 and protein substrate (3, 30, 31) .
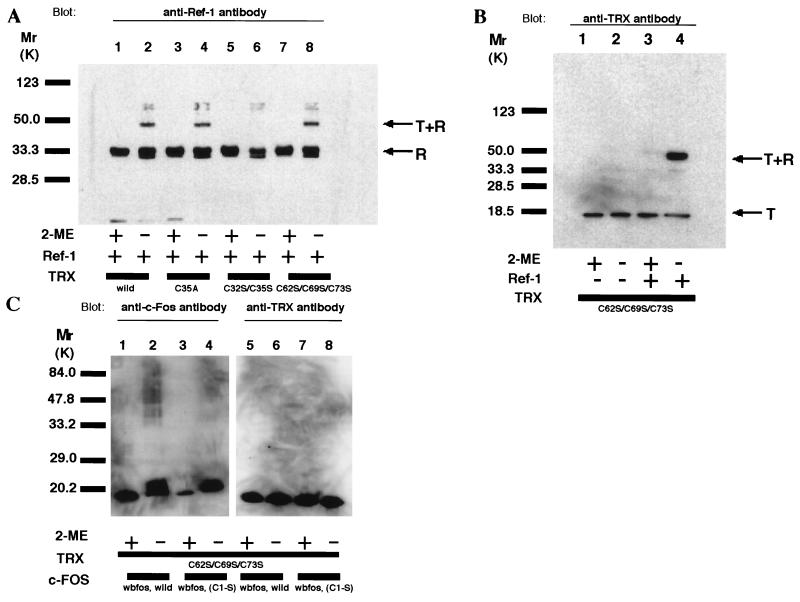
Under reducing conditions, the wild-type TRX and all the TRX mutants (TRXC32S/C35S, TRXC35A, and TRXC62S/C69S/C73S) migrated as a single 13-kDa band in SDS/PAGE. Treatment of TRXwild or TRXC32S/C35S with diamide generated multiple bands, suggesting oligomer formation. In the case of TRXC62S/C69S/C73S, however, we detected only a monomeric form after diamide treatment (data not shown). Ref-1 has seven cysteines, and it moved as a monomer under reducing conditions (Fig. 4A, lanes 1, 3, 5, and 7). In oxidizing conditions, an additional band, migrating faster than the reduced form, was observed; this probably was a different monomeric form of Ref-1, with intramolecular disulfide bond (Fig. 4A, lanes 2, 4, 6, and 8) (18). We found no additional bands of higher molecular size, suggesting that Ref-1 did not form cysteine-mediated multimers under the thiol-targeted oxidizing conditions. Next, wild-type or TRX mutants were incubated with Ref-1, and then their migration patterns were analyzed. Both antibodies against Ref-1 (Fig. 4A) and TRX (Fig. 4B) recognized the same extra bands migrating around 50 kDa in lanes of TRXwild, TRXC35A, and TRXC62S/C69S/C73S (Fig. 4A, lanes 2, 4, and 8; Fig. 4B, lane 4) under oxidizing conditions, but not of TRXC32S/C35S species (Fig. 4A, lane 6). Only TRXC62S/C69S/C73S was presented in Fig. 4 B and C because the other TRXs (wild, C35A, C32S/C35S) form multimers by themselves, which are difficult to discriminate from heterocomplexes.
Figure 4.
Evidence that TRX associated with Ref-1 through the redox-active cysteines in vitro. Various TRX mutants containing cysteine-to-serine or cysteine-to-alanine substitutions were cross-linked with Ref-1 or wbFos by diamide. After incubation, the complexes were resolved by electrophoresis under reducing or oxidizing conditions and detected by antibodies. (A) Western blot analysis of various TRX and Ref-1 complex by use of anti-Ref-1 antibody. (B) Western blot analysis of TRXC32S/C69S/C73S and Ref-1 complex by use of anti-TRX antibody. (C) Western blot analysis of TRXC32S/C69S/C73 and wbFos by use of anti-c-Fos antibody or anti-TRX antibody.
To exclude the possibility that diamide could cross-link any cysteine-containing proteins nonspecifically, c-Fos–TRX interaction was assayed in the same condition. c-Fos, which also has a well-conserved cysteine in its DNA-binding domain (DBD), is one of the redox-sensitive transcription factors and is known to be controlled by Ref-1. Fos polypeptides, wbFos and wbFos (C1-S), were incubated with or without TRXC62S/C69S/C73S. As shown in Fig. 4C, we could not detect covalent dimers either between wbFos themselves or with TRXC62S/C69S/C73S.
These results showed that TRX and Ref-1 preferentially formed a covalent heterodimer in vitro and further suggested that this association involved Cys-32 in the active center of TRX.
Demonstration of a Physical Association Between TRX and Ref-1 by the Mammalian Two-Hybrid Assay in COS-7 Cells.
We investigated if TRX could associate with Ref-1 in the nucleus using the two-hybrid system with cultured mammalian cells (22). A complementary DNA of TRX or Ref-1 was subcloned downstream of the DBD of GAL4 (23) or the VP16 transactivation domain (24). These fusion plasmids were then coexpressed in COS-7 cells with a plasmid containing the luciferase gene as a reporter.
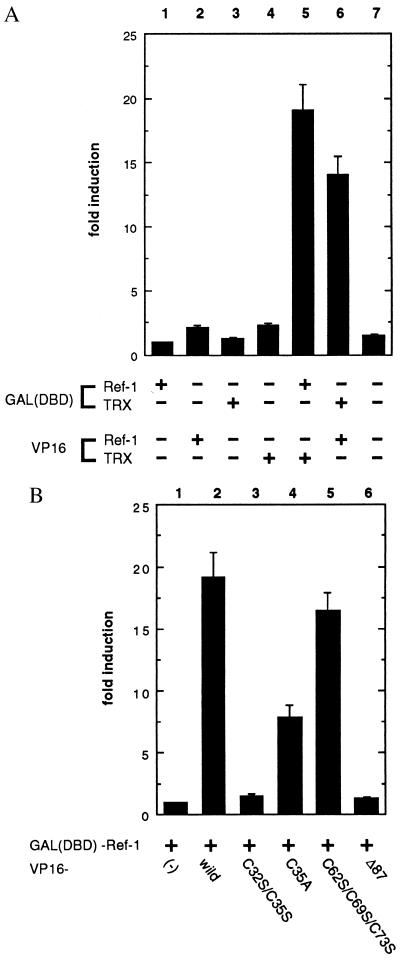
As shown in Fig. 5A, when a DBD–TRX or DBD–Ref-1 was expressed in COS-7 cells with VP16, there was no luciferase activity (lanes 1 and 3). When VP16–TRX or VP16–Ref-1 was expressed with DBD, only marginal inductions were detected (lanes 2 and 4). In contrast, coexpression of the wild-type TRX and Ref-1 fusion constructs induced the luciferase activity more than 10-fold (lanes 5 and 6) over the basal level (lane 7). Next, we used TRX mutants to identify the cysteine residue(s) responsible for this interaction (Fig. 5B). A triple mutation of the noncatalytic cysteines (Cys-62, Cys-69, and Cys-73) did not diminish the luciferase activity (lane 5), suggesting that these cysteines are dispensable for the activity of TRX to interact with Ref-1. Compatible with our in vitro experiments, significant luciferase induction was observed by coexpression of TRXC35A and Ref-1 fusion constructs (lane 4). On the other hand, coexpression of TRXC32S/C35S and Ref-1 fusion constructs failed to induce luciferase activity (lane 3). These in vitro and in vivo data showed that the direct physical association between TRX and Ref-1 was mediated through the active center of TRX. In addition to substitution mutants, we constructed a truncated TRX, TRXΔ87 (Fig. 1) by deleting the C-terminal domain from amino acid 88. TRXΔ87 has all its five cysteines preserved but has lost its dithiol reducing activity. Coexpression of TRXΔ87 and Ref-1 fusion constructs showed no significant induction of luciferase activity (lane 6).
Figure 5.
Demonstration of a physical association between TRX and Ref-1 by mammalian two-hybrid assay in COS-7 cells. (A) An association between wild-type TRX and Ref-1 was monitored by a luciferase assay. (B) An association between TRX mutants and Ref-1 was monitored by a luciferase assay. The relative fold induction of luciferase activity was calculated by normalizing to the β-galactosidase activity. The results are the means ± SE of three experiments (each in duplicate) and presented as fold increases in luciferase activity over the baseline seen in lane 1.
DISCUSSION
Redox regulation of transcription factors is an interesting and important issue. The result of in vitro gel shift assays has established that the DNA-binding activity of a growing number of transcription factors, including AP-1, NF-κB, Myb, and Ets, is regulated by a thiol-redox control mechanism (8, 32–35). Although some chemical agents, such as 2-mercaptoethanol and DTT, can activate DNA binding of transcription factors, the issue of which endogenous molecules act as physiological redox regulators is still to be clarified. In the case of NF-κB, TRX is one of the candidate molecules: Qin et al. (30) demonstrated direct physical association between TRX and an oligopeptide from the DNA-binding loop of NF-κB p50. That and previous reports strongly suggest that NF-κB is a target molecule of redox regulation by TRX (8, 9, 36).
As a representative model of redox regulation of transcription factors, the molecular mechanism of AP-1 redox regulation has been intensively studied in vivo. Xanthoudakis et al. (19) showed that AP-1 was not a direct substrate of TRX. Because the amino acid sequence surrounding the cysteine is entirely different from that of NF-κB (37, 38), not surprisingly TRX alone is unable to activate AP-1 DNA binding by cysteine reduction in vitro. Moreover, in the case of AP-1, interestingly redox regulation of AP-1 involves a conserved cysteine residue in the DBD (1, 32). Substitution of the cysteine with a serine results in a gain-of-function phenotype (37); this substitution occurs spontaneously during the generation of the v-jun oncogene and may contribute to v-jun’s oncogenic properties (38, 39). Analysis of mutated fos genes in cell transformation assays suggests that redox regulation also affects Fos activity in vivo via these conserved cysteines. These reports show that modification of redox-sensitive cysteines of the AP-1 molecules may control its transcriptional activity and regulate subsequent gene inductions. These types of substitutions in NF-κB bring complete loss of function (8). Thus, redox regulation of AP-1 seems necessary for appropriate gene expression.
Ref-1 was cloned as a molecule that stimulated DNA binding activity of AP-1 (19). However, no report has described the electron donors for Ref-1 molecules. In this paper, we have shown that TRX is one of the endogenous redox molecules and regulates the AP-1 transcriptional activity through its direct association with Ref-1. In other words, TRX is one of the hydrogen donors of Ref-1. Indeed, Qin et al. (40) solved the solution structure of an oligopeptide comprising amino acid residues 59–71 of Ref-1 and TRX complex, thus demonstrating the direct association of TRX and the partial peptide of Ref-1. Our in vitro data, using whole recombinant molecules, and in vivo experimental results show that Cys-32 and Cys-35 of TRX, which constitute the catalytic center, are involved in the association. Site-directed mutagenesis studies showed that two cysteines of the redox domain of Ref-1, Cys-63 and Cys-95, are redox sensitive (41, 42) and can be targets of TRX.
As a way of transcriptional control, the subcellular locations of some transcription factors are strictly regulated. One of the best examples is a nuclear translocation of NF-κB, which has a nuclear localization signal sequence (NLS) under activation stimuli (43, 44). In contrast, TRX has no authentic NLS. Although the minute translocational mechanism is to be investigated in a further study, we have observed that UV irradiation, which also potentiates the AP-1 activity, translocates TRX into the nucleus in the HSC-1 keratinocyte cell line and HeLa cells (45). As shown in Fig. 2, overexpression of TRX and Ref-1 alone was not sufficient to induce significant luciferase expression, and this result suggests that nuclear translocation of TRX is indispensable for potentiating the AP-1 transcriptional activity. These data collectively suggest that PMA translocates TRX into the nucleus, allowing its interaction with Ref-1, and that this association promotes the direct activation of AP-1 by Ref-1. Recently, nuclear translocation of mitogen-activating protein kinases on stimulation has been implicated (46, 47). Interestingly, redox-acting molecules as well as kinases regulate transcription activity in the nucleus.
We failed to immunoprecipitate Ref-1 with the anti-TRX antibody from HeLa cells. If TRX is a hydrogen donor for Ref-1, their association would be transient and too weak to be detectable by standard immunoprecipitation methods. To get higher sensitivity for detection of an association between TRX and Ref-1, we used a diamide-induce cross-linking assay and the in vivo two-hybrid assay. Considering that diamide is a potent sulfhydryl-oxidizing agent, it should be able to cross-link various proteins nonspecifically, depending on the assay conditions. However, this is not the case in our experiments because c-Fos and TRXC62S/C69S/C73S did not form such a complex in the same condition. The mammalian two-hybrid assay has been used to study various protein–protein interactions, and it is especially useful in confirming interactions identified by two-hybrid library screens in yeast. This assay can also be used in combination with deletional or site-directed mutagenesis to rapidly map the amino acids responsible for the interaction between two proteins. In the two-hybrid assay, reporter gene expressions imply the association of two fusion proteins in living cell nuclei. Accordingly, in this report, the in vitro cross-linking assay and the mammalian two-hybrid assay were complementary in proving the physiological association of TRX and Ref-1. Our results showed that TRX associates with Ref-1 via the catalytic center of TRX in living cells. Coexpression of TRX lacking a C-terminal domain and Ref-1 fusion constructs showed no significant induction of luciferase activity; this result suggests that the C-terminal region of TRX is indispensable for its interaction with Ref-1 as well as for its reducing activity.
In the present study, we demonstrated the PMA-dependent translocation of cytoplasmic TRX to the nuclear compartment and the direct physical association between TRX and Ref-1 in vitro and in vivo. The precise mechanism and the role of TRX–Ref-1 interaction in the regulation of gene expression is to be clarified.
Acknowledgments
We thank S. Xanthoudakis and K. Umesono for plasmid vectors, T. Curran for useful consultation, S. Toyama and H. Kagoshima for technical advice, W. Brown and R. Yamaguchi for review of the manuscript, G. M. Clore and A. Gronenborn for communicating their results before publications, and Y. Kanekiyo and C. Ogawa for secretarial help. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan.
ABBREVIATIONS
- TRX
thioredoxin
- NF
nuclear factor
- AP-1
activator protein-1
- Ref-1
redox factor-1
- PMA
phorbol 12-myristate 13-acetate
- 6xHis
hexahistidine
- DBD
DNA-binding domain
References
- 1.Pahl H L, Baeuerle P A. BioEssays. 1994;16:497–502. doi: 10.1002/bies.950160709. [DOI] [PubMed] [Google Scholar]
- 2.Laurent T C, Moore E C, Reichard P. J Biol Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 3.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 4.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 5.Buchanan B B, Schurmann P, Decottignies P, Lozano R M. Arch Biochem Biophys. 1994;314:257–260. doi: 10.1006/abbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- 6.Tagaya Y, Maeda Y, Mitsui A, Kondo N, Masutani H, Hamuro J, Brown N, Arai K, Yokota T, Wakasugi H, Yodoi J. EMBO J. 1989;8:757–764. doi: 10.1002/j.1460-2075.1989.tb03436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yodoi J, Uchiyama T. Immunol Today. 1992;13:405–411. doi: 10.1016/0167-5699(92)90091-K. [DOI] [PubMed] [Google Scholar]
- 8.Matthews J R, Wakasugi N, Virelizier J L, Yodoi J, Hay R T. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto T, Ogiwara H, Hayashi T, Mitsui A, Kawabe T, Yodoi J. Int Immunol. 1992;4:811–819. doi: 10.1093/intimm/4.7.811. [DOI] [PubMed] [Google Scholar]
- 10.Sasada T, Iwata S, Sato S, Kitaoka T, Hirota K, Nakamura K, Nishiyama A, Y, Taniguchi Y, Takabayashi A, Yodoi J. J Clin Invest. 1996;97:2268–2276. doi: 10.1172/JCI118668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silberstein D S, McDonough S, Minkoff M S, Balcewicz Sablinska M K. J Biol Chem. 1993;268:9138–9142. [PubMed] [Google Scholar]
- 12.Biguet C, Wakasugi N, Mishal Z, Holmgren A, Chouaib S, Tursz T, Wakasugi H. J Biol Chem. 1994;269:28865–28870. [PubMed] [Google Scholar]
- 13.Wakasugi N, Tagaya Y, Wakasugi H, Mitsui A, Maeda M, Yodoi J, Tursz T. Proc Natl Acad Sci USA. 1990;87:8282–8286. doi: 10.1073/pnas.87.21.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Clore G M, Gronenborn A M. Structure (London) 1994;2:503–522. doi: 10.1016/s0969-2126(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 15.Meyer M, Schreck R, Baeuerle P A. EMBO J. 1993;12:2005–2015. doi: 10.1002/j.1460-2075.1993.tb05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenk H, Klein M, Erdbrügger W, Dröge W, Schulze-Osthoff K. Proc Natl Acad Sci USA. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xanthoudakis S, Curran T. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xanthoudakis S, Miao G G, Curran T. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xanthoudakis S, Miao G, Wang F, Pan Y C, Curran T. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 21.Angel P, Baumann I, Stein B, Delius H, Rahmsdorf H J, Herrlich P. Mol Cell Biol. 1987;7:2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nerlov C, Ziff E B. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadowski I, Ptashne M. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlmann T, Rangarajan P N, Umesono K, Evans R M. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 25.Barzilay G, Hickson I D. BioEssays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 26.Isoda K, Nüsslein-Volhard C. Proc Natl Acad Sci USA. 1994;91:5350–5354. doi: 10.1073/pnas.91.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishigami S, Kannaya E, Kikuchi M, Ito K. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 28.Oblong J E, Berggren M, Gasdaska P Y, Powis G. J Biol Chem. 1994;269:11714–11720. [PubMed] [Google Scholar]
- 29.Tonissen K, Wells J, Cock I, Perkins A, Orozco C, Clarke F. J Biol Chem. 1993;268:22485–22489. [PubMed] [Google Scholar]
- 30.Qin J, Clore G M, Kennedy W M P, Huth J R, Gronenborn A M. Structure (London) 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 31.Forman-Kay J D, Clore G M, Gronenborn A M. Biochemistry. 1992;31:3442–3452. doi: 10.1021/bi00128a019. [DOI] [PubMed] [Google Scholar]
- 32.Abate C, Patel L, Rauscher F J d, Curran T. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 33.Wasylyk C, Wasylyk B. Nucleic Acids Res. 1993;21:523–529. doi: 10.1093/nar/21.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myrset A H, Bostad A, Nadege J, Pierre-Noel L, Toma F, Gabrielsen O S. EMBO J. 1993;12:4625–4633. doi: 10.1002/j.1460-2075.1993.tb06151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guehmann S, Vorbruehggen G, Kalkbrenner F, Moelling K. Nucleic Acids Res. 1992;20:2279–2286. doi: 10.1093/nar/20.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi T, Ueno Y, Okamoto T. J Biol Chem. 1993;268:11380–11388. [PubMed] [Google Scholar]
- 37.Okuno H, Akahori A, Sato H, Xanthoudakis S, Curran T, Iba H. Oncogene. 1993;8:695–701. [PubMed] [Google Scholar]
- 38.Ng L, Forrest D, Curran T. Nucleic Acids Res. 1993;21:5831–5837. doi: 10.1093/nar/21.25.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chida K, Vogt P K. Proc Natl Acad Sci USA. 1992;89:4290–4294. doi: 10.1073/pnas.89.10.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J, Clore G M, Kennedy W M P, Kuszewski J, Gronenborn A M. Structure (London) 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- 41.Walker L J, Robson C N, Black E, Gillespie D, Hickson I D. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker L J, Craig R B, Harris A L, Hickson I D. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henkel T, Zabel U, Zee K V, Müller J M, Fanning E, Baeuerle P A. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 44.Yoneda Y, Imamoto-Sonobe N, Matsuoka Y, Iwamoto R, Kiho Y, Uchida T. Science. 1988;242:275–278. doi: 10.1126/science.3051382. [DOI] [PubMed] [Google Scholar]
- 45.Masutani, H., Hirota., K., Sasada, T., Ueda-Taniguchi, Y., Taniguchi, Y., Matsui, M. & Yodoi, J., Immunol. Lett., in press. [DOI] [PubMed]
- 46.Tyrrell R M. BioEssays. 1995;18:139–148. doi: 10.1002/bies.950180210. [DOI] [PubMed] [Google Scholar]
- 47.Karin M, Hunter T. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]