Abstract
Protein homeostasis maintains proper intracellular balance by promoting protein folding and clearance mechanisms while minimizing the stress caused by the accumulation of misfolded and damaged proteins. Chronic expression of aggregation-prone proteins is deleterious to the cell and has been linked to a wide range of conformational disorders. The molecular response to misfolded proteins is highly conserved and generally studied as a cell-autonomous process. Here, we provide evidence that neuronal signaling is an important modulator of protein homeostasis in post-synaptic muscle cells. In a forward genetic screen in Caenorhabditis elegans for enhancers of polyglutamine aggregation in muscle cells, we identified unc-30, a neuron-specific transcription factor that regulates the synthesis of the inhibitory neurotransmitter γ-aminobutyric acid (GABA). We used additional sensors of protein conformational states to show that defective GABA signaling or increased acetylcholine (ACh) signaling causes a general imbalance in protein homeostasis in post-synaptic muscle cells. Moreover, exposure to GABA antagonists or ACh agonists has a similar effect, which reveals that toxins that act at the neuromuscular junction are potent modifiers of protein conformational disorders. These results demonstrate the importance of intercellular communication in intracellular homeostasis.
[Keywords: Protein misfolding, polyglutamine aggregation, GABAergic neurons, cholinergic neurons, protein polymorphisms]
All organisms have in common the necessity to maintain intracellular homeostasis, a balance between protein synthesis, folding, and clearance, and the ability to respond to environmental challenges and pathophysiological conditions (Dobson 2003). Protein quality control mechanisms prevent the appearance of misfolded and aggregation-prone proteins. Disruption in protein homeostasis leads to the appearance and accumulation of intermediate nonnative conformations that tend to form oligomeric and aggregated species, which over time cause cellular injury (Soto 2003; Gregersen 2006). This problem is fundamental to cell biology, and is associated with an increasing number of diseases of protein conformation (Soto 2003; Brignull et al. 2007). For each disorder, the expression of a specific mutant protein can result in either loss of function—for example, cystic fibrosis, in which there is premature clearance of the misfolded mutant protein (Amaral 2006; Gregersen 2006)—or toxic gain of function such as Huntington’s disease or amyotrophic lateral sclerosis (ALS), where misfolded mutant proteins accumulate and cause toxicity (Cohen and Kelly 2003). Diseases associated with proteotoxicity have been suggested to share common molecular and cellular pathogenic mechanisms (Dobson 2003; Stefani 2004; Drysdale et al. 2006; Rusmini et al. 2007).
The genetic background of an organism and its interaction with the environment are likely contributors to the network of regulators that impact on the course of conformational disorders. The eukaryotic proteome has abundant polymorphisms (Kimchi-Sarfaty et al. 2007) that are maintained in a phenotypically silent state or can become exposed by environmental fluctuations or changes in the folding capacity of the cell (Rutherford and Lindquist 1998; Queitsch et al. 2002; Gidalevitz et al. 2006). For example, the expression of mutant polyglutamine (polyQ) expansion proteins causes the global loss of function of other coexpressed metastable polymorphic proteins, and moreover, the “unmasking” of these metastable polymorphisms further enhances polyQ misfolding and aggregation (Gidalevitz et al. 2006). These results suggest that endogenous protein polymorphisms can act as genetic modifiers in protein conformational diseases. This may be relevant to other late-onset progressive neurodegenerative diseases such as ALS, for which 90% of the cases are sporadic, but share the same pathological characteristics of the familial form of the disease (Boillee et al. 2006).
Proteins containing expanded polyQ form alternate conformational states that can self-associate and form proteotoxic oligomeric and aggregate species (Warrick et al. 1998; Gusella and MacDonald 2000; Chen et al. 2002). For polyQ repeat diseases, the length of the polyQ expansion dictates both the kinetics of aggregation (Chen et al. 2002) and age of disease onset (Gusella and MacDonald 2000). A number of Caenorhabditis elegans models expressing polyQ proteins have been established in which length-dependent phenotypes have been observed in different tissues (Faber et al. 1999; Parker et al. 2001). The C. elegans models established by our laboratory for polyQ aggregation and toxicity employed chimeric polyQ fusion proteins to yellow fluorescent protein (YFP) that revealed a clear length- and age-dependent polyQ aggregation and cellular toxicity in body wall muscle cells (Satyal et al. 2000; Morley et al. 2002) and neurons (Brignull et al. 2006) that mimics the polyQ-length dependence observed for Huntington’s disease. Animals expressing 24 glutamines (Q24) exhibited soluble fluorescence, whereas animals expressing 35 (Q35) or 40 glutamines (Q40) exhibited age-dependent aggregation (Nollen et al. 2004). The intrinsic characteristics of polyQ protein aggregation and toxicity in C. elegans have proven useful for genome-wide screens for modifiers of protein aggregation (Nollen et al. 2004) and as probes to challenge the folding environment revealing the delicate nature of protein homeostasis (Nollen et al. 2004; Gidalevitz et al. 2006). These models also contributed to establish the critical roles of the age-1/daf-2 insulin-like signaling (ILS) signaling pathways and the heat-shock response as potent modulators of polyQ aggregation and toxicity and as lifespan regulators (Morley et al. 2002; Hsu et al. 2003; Nollen et al. 2004). Collectively, these studies revealed the delicate nature of protein homeostasis, the relative ease by which an imbalance occurs and the role of stress signaling networks of intracellular factors to counter this imbalance.
In this study, we show that neuronal signaling is a potent non-cell-autonomous modulator of post-synaptic protein homeostasis. Disturbances in neuronal signaling, whether caused by genetic modifications or pharmacological agents that lead to post-synaptic cellular overstimulation, have widespread effects on the cell’s buffering capacity for protein folding resulting in misfolding of conformationally challenged metastable proteins. Therefore, changes in cell–cell communication represent an important and previously unidentified modulator of protein homeostasis. Furthermore, due to the ability of cell–cell communication to affect protein folding, non-cell-autonomous regulation is likely to represent a critical component of protein misfolding disorders.
Results
rm7 leads to premature polyQ aggregation
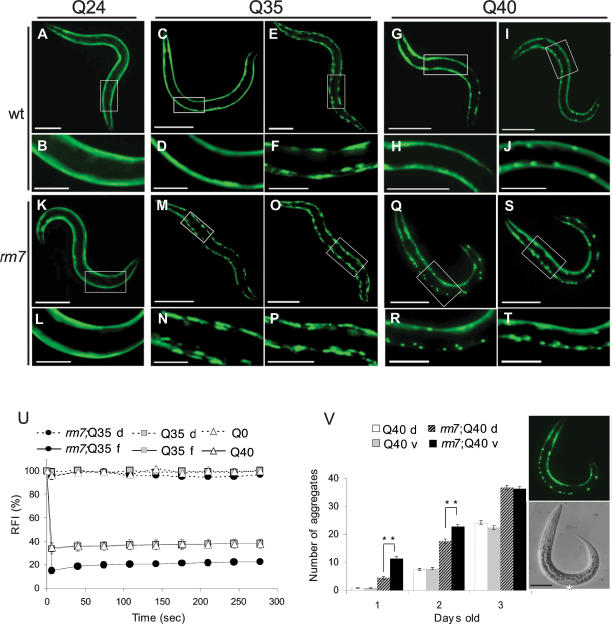
We identified novel modifiers of polyQ aggregation by performing a forward genetic screen of C. elegans strains expressing polyQ∷YFP in body wall muscle cells (Fig. 1A–J). Mutants that exhibited a premature shift from a diffuse to punctate polyQ∷YFP distribution were selected. We identified and characterized a mutation, rm7, that led to the premature appearance of polyQ foci in Q35∷YFP or Q40∷YFP animals compared with wild-type Q35 and Q40 lines, respectively (Fig. 1, A–J,K–T). In Q35;rm7 animals, the polyQ foci appeared prematurely in young adult (4-d-old) animals and increased as the animals aged (Fig. 1M–P). By comparison, in wild-type Q35 animals polyQ foci appeared initially at a later age (5 d after hatching), with widespread distribution only at 6 d of age (Fig. 1C–F). Likewise, premature polyQ foci were also observed in rm7;Q40 animals as young day 1 larvae (Fig. 1Q,R), a full day earlier than for wild-type Q40 animals (Fig. 1G–J). In 1-d-old rm7;Q40 animals the appearance of polyQ foci displayed a striking dorsal–ventral asymmetry, with a prevalence of foci on the ventral side (Fig. 1Q–T,V). This dorsal–ventral asymmetry in distribution was only observed early in development (Fig. 1V). The rm7 mutation had no effect in Q24-expressing animals (Fig. 1A,B,K,L).
Figure 1.
The mutation rm7 causes premature polyQ aggregation in C. elegans expressing Q35∷YFP or Q40∷YFP in body wall muscle cells. Fluorescent microscopy images of 6-d-old Q24 (A,B) and rm7;Q24 (K,L), 4-d-old Q35 (C,D) and Q35;rm7 (M,N), 6-d-old Q35 (E,F) and Q35;rm7 (O,P), 1-d-old Q40 (G,H) and rm7;Q40 (Q,R), and 2-d-old Q40 (I,J) and rm7;Q40 (S,T) animals. The white box indicates magnified region. Bars: A,C,E,K,M,O, 200 μm; B,D,F,G,I,L,N,P,Q,S, 50 μm; H,J,R,T, 25 μm. (U) Quantitative FRAP analysis of diffuse or foci Q35∷YFP proteins. Full lines represent “foci” and dashed lines represent “diffuse protein,” indicated by f and d, respectively. The relative fluorescence intensity (RFI) was determined for each time point and represents the average of a minimum of four independent measurements. Error bars indicate SEM. (V) Dorsal–ventral aggregation asymmetry. Aggregates in the dorsal and ventral sides of Q40 and rm7;Q40 age-matched animals were quantified. Columns represent the number of aggregates in the dorsal side of Q40 (white columns), ventral side of Q40 (gray columns), dorsal side of rm7;Q40 (dashed columns), and ventral side of rm7;Q40 (black columns) animals. In the figure, d and v indicate dorsal and ventral, respectively. Each column represents the average of 80 animals and error bars indicate SEM. (**) P < 0.01 between dorsal and ventral rm7;Q40. Fluorescent and phase-contrast images of 1-d-old rm7;Q40 animals. White asterisk indicates the region were vulva will form at a late larval stage corresponding to the ventral side of the animal. Bar, 50 μm.
We next examined whether the polyQ foci detected in the rm7 background corresponded to the appearance of immobile aggregate species using fluorescence recovery after photobleaching (FRAP) analysis. FRAP is a dynamic imaging method that we adapted to cells of living worms to assess changes in protein mobility and solubility (Brignull et al. 2006). The foci in day 4 Q35;rm7 animals did not recover following photobleaching (Fig. 1U), indicating that that Q35 had changed from a soluble, diffuse population to an immobile and aggregated state. These results suggest that the rm7 mutation leads to the premature appearance of polyQ aggregates. Similar results were also observed for the polyQ foci formed in rm7;Q40 animals (data not shown) in contrast to soluble Q35 or Q40 in wild-type backgrounds. We concluded that the rm7 mutation caused the premature conversion of expanded polyQ∷YFP from a soluble to an aggregated state in body wall muscle cells. We addressed whether the rm7 mutation altered the levels of polyQ protein by Western blot analysis and found that wild-type and rm7 animals express equivalent levels of Q35 (Supplementary Fig. S1). These results reveal that the premature polyQ aggregation observed in the rm7 background is not due to changes in polyQ protein levels, and rather is due to an imbalance in protein homeostasis.
rm7 is a hypomorphic allele of unc-30
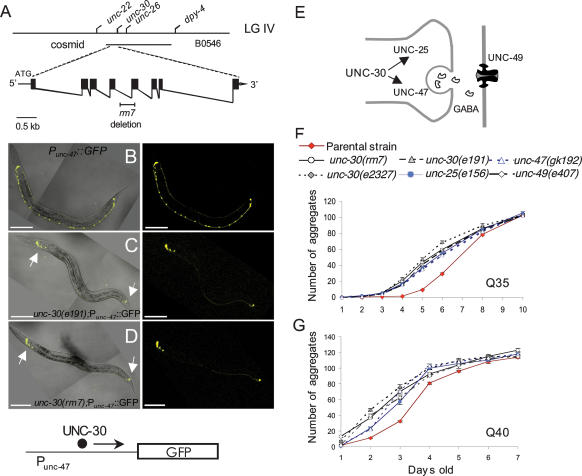
Using standard genetic techniques, the rm7 mutation was mapped to the gene unc-30 (Fig. 2A; Supplementary Fig. S2; Supplemental Material). UNC-30 is a neuron-specific homeodomain transcription factor essential for γ-aminobutyric acid (GABA) synthesis (Jin et al. 1994). GABAergic motor neurons are required for proper locomotion and both unc-30-null alleles and unc-30(rm7) cause an obvious uncoordinated phenotype (Supplemental Material). The rm7 mutation was characterized and shown to be due to a 347-base-pair (bp) deletion of part of the homeodomain region, the DNA-binding domain of UNC-30 (Supplementary Fig. S3). Consequently, the phenotype of the rm7 mutation has an uncoordinated phenotype indistinguishable from that of unc-30-null alleles (Supplemental Material).
Figure 2.
GABAergic-defective signaling leads to premature aggregation of polyQ expansions. (A) Above is the genetic map of the unc-30 region on chromosome IV and rescuing cosmid B0564; the bottom displays the unc-30 exon–intron structure containing the rm7 deletion region. (B–D) Fluorescent images and merged phase-contrast images of young adult animals Punc-47∷gfp (B), unc-30(e191);Punc-47∷gfp (C), and unc-30(rm7);Punc-47∷gfp (D). Animals expressing Punc-47∷gfp exhibit GFP expression in the 26 GABAergic neurons (B); conversely, unc-30(e191);Punc-47∷gfp (C), and unc-30(rm7);Punc-47∷gfp (D) display GFP localization only in the non-type-D GABAergic motor neurons (white arrows). Bar, 100 μm. (E) Schematic representation of the downstream targets of UNC-30. (F,G) Animals were synchronized and the age-dependent accumulation of aggregates was determined. The strains analyzed were Q35 (red diamonds), Q35;unc-30(rm7) (empty circles), Q35;unc-30(e191) (gray triangles), Q35;unc-30(e2327) (gray diamonds), Q35;unc-25(e156) (blue circles), and Q35;unc-47(gk192) (empty blue triangles) (F); and Q40 (red diamonds), unc-30(rm7);Q40 (empty circles), unc-30(e2327);Q40 (gray diamonds), unc-25(e156);Q40 (blue circles), unc-47(gk192);Q40 (empty blue triangles), and unc-49(e407);Q40 (empty gray diamonds) (G). Cohort sizes decreased as some animals died during the experiment, but each data point represents at least 21 animals (range, 21–141). Error bars indicate SEM.
The effect of the rm7 mutation on UNC-30 activity and GABA signaling was examined using a reporter strain containing the promoter region of unc-47, the vesicular GABA transporter that is a downstream target of UNC-30 (Fig. 2E; McIntire et al. 1997). The unc-47 promoter fused to GFP, Punc-47∷gfp, is expressed in all 26 GABAergic neurons (Fig. 2B; McIntire et al. 1997). Introduction of an unc-30-null mutation into the background of the reporter strain, unc-30(e191);Punc-47∷gfp, results in a complete absence of GFP expression in the 19 type-D GABAergic neurons with no effect on seven, UNC-30-independent, non-type-D GABAergic neurons (Fig. 2C). A similar loss of GFP expression was observed for unc-30(rm7);Punc-47∷GFP in the 19 type-D GABAergic neurons (Fig. 2D). We concluded from these results that the unc-30(rm7) allele is a hypomorphic allele of unc-30 that results in a severe deficit in GABAergic signaling.
Defective GABAergic signaling leads to premature polyQ aggregation
A prediction from the unc-30(rm7) result is that other unc-30 alleles as well as mutations in UNC-30 downstream targets should also result in premature polyQ aggregation. To test this, we examined two known unc-30-null alleles and mutations in two UNC-30 targets, unc-47 encoding vesicular GABA transporter and unc-25, the gene encoding glutamic acid decarboxylase (Fig. 2E; Jin et al. 1999). All of these mutations resulted in the premature appearance of polyQ aggregation, similar to what we had observed for unc-30(rm7) (Fig. 2F,G). On average, at day 4, mutants in the UNC-30 pathway had a 17-fold increase in polyQ35 aggregation with respect to Q35 wild-type animals. This increase in aggregation persisted through day 5, when GABA-defective mutant animals displayed a fourfold increase in aggregates relative to wild-type animals. Furthermore, Q35 GABA-defective animals have an enhanced aggregation half-time (t1/2, defined as the average age at which animals reached 50% maximal aggregation) of 1.2 d earlier than Q35 wild-type control animals (Supplementary Table S1). Q40 GABA-defective animals in turn displayed a threefold increase in aggregation at day 2 and a nearly 1-d enhancement in the t1/2 (Supplementary Table S1). We next examined animals with a null mutation for unc-49 encoding the GABA receptor expressed in body wall muscle cells (Fig. 2E). unc-49(e407);Q40 animals also exhibited premature Q40∷YFP aggregation, similar to that observed in other GABA mutants (Fig. 2G). These results revealed a consistent pattern of premature polyQ aggregation and suggested that defective GABA signaling in motor neurons affected protein aggregation in post-synaptic muscle cells.
Additional support for the involvement of GABAergic signaling on post-synaptic protein homeostasis is provided by our earlier observation of asymmetric dorsoventral pattern of Q40 aggregation in unc-30 animals (Fig. 1Q–T,V). The timing of Q40 dorsal–ventral aggregate distribution correlates with the asymmetric development of GABAergic neuromuscular innervation. In L1 larval stage animals, GABAergic signaling is present only in the ventral body wall muscle cells and by L2, both dorsal and ventral sides become innervated (White et al. 1978). The initial cells that are exposed to defective GABA signaling are the ventral body wall muscle cells. We found a striking correlation with the appearance of aggregates predominately on this side (Fig. 1V). Furthermore, this asymmetry in aggregation onset was observed for the different Q40;GABA-defective strains analyzed (data not shown). As expected, this dorsal–ventral asymmetry in polyQ aggregation was transient and was not observed later in development (Fig. 1V) at 3 d old. Taken together, these results provide strong support to a model in which premature polyQ aggregation is induced by defective GABAergic signaling in the post-synaptic cell.
Cholinergic hyperstimulation causes premature polyQ aggregation in body wall muscle cells
The sinusoidal locomotion of C. elegans results from alternate body wall muscle contraction, promoted by excitatory cholinergic motorneurons, and muscle relaxation promoted by inhibitory GABAergic motorneurons. We hypothesized that the premature polyQ aggregation observed in GABA-defective mutants was the result of overstimulation of the post-synaptic cells by the cholinergic motorneurons. To test this, we analyzed the effects of mutations in acetylcholinesterases ace-1 and ace-2 that result in muscle hypercontraction due to the reduction of acetylcholine (ACh) clearance from the neuromuscular junction (NMJ) (Combes et al. 2003). In Q35;ace-2(g72);ace-1(p1000) mutant animals, we observed a premature enhancement (more than twofold) in Q35∷YFP aggregation at 4 d relative to controls (Fig. 3A). These results indicate that increased cholinergic activity results in premature polyQ aggregation.
Figure 3.
Cholinergic hyperstimulation causes premature polyQ aggregation in body wall muscle cells. Animals were synchronized and the age-dependent accumulation of aggregates was determined by fluorescent microscopy. (A,B) The strains analyzed were Q35, Q35,ace-2(g72);ace-1(p1000) (A) and Q35, Q35;egl-19(ad695) (B). Each column represents a minimum of 20 animals (range of 20–72). (C–F) One-day-old (L1) animals were treated with 200 mM GABA (C), 1 mM lindane (D), 25 μM levamisole (E), or 1 mM nicotine (F), and the number of aggregates were determined daily. (C–F) The age-dependent increase in aggregate number was plotted for the different drug treatments. An n value of at least 40 animals (range, 40–200) for each concentration and time point was analyzed. For all graphs, error bars indicate SEM. (*) P < 0.05; (**) P < 0.01.
We next examined the role of downstream components of ACh signaling. The pathway activated downstream from the ACh receptor is calcium (Ca2+) dependent (Richmond and Jorgensen 1999; Jospin et al. 2002); therefore, an increase of Ca2+ influx should result in premature polyQ aggregation. To address this, we employed a gain-of-function mutation in the α1-subunit of the Ca2+ channel, egl-19(ad695), shown to yield a sustained influx of Ca2+ into body wall muscle cells (Lee et al. 1997). The aggregation phenotype of Q35;egl-19(ad695) animals displayed a dramatic 16-fold increase in aggregation at day 4 that corresponds to a 1-d enhancement in t1/2 (Fig. 3B; Supplementary Table S1), with a premature aggregation onset similar to the observed for Q35 GABA-defective animals (Fig. 2F). Conversely, a partial loss-of-function mutation in this gene, egl-19(n582), leading to a 30% decrease in the amount of Ca2+ entering the muscle cells, caused a decrease in aggregation in Q35;egl-19(n582) animals (Supplementary Fig. S4A). Together, these data indicate that overexcitation of post-synaptic muscle cells is the basis for an imbalance in protein homeostasis. This hypothesis was further corroborated by the finding that a reduction in the general excitability of body wall muscle cells can also reduce polyQ aggregation. Specifically, we observed that a gain-of-function mutation in unc-93, a gene encoding a subunit of the K+ channel complex that is known to reduce muscle contraction (de la Cruz et al. 2003), decreased polyQ aggregation in body wall muscle cells (Supplementary Fig. S4B). Collectively, these results suggest that a shift in the signaling balance toward cellular overexcitation underlies the disruption in protein homeostasis and lend support for the role of neuronal signaling as an important modulator of polyQ misfolding and aggregation.
Small molecule agonists and antagonists that alter GABAergic and cholinergic signaling affect protein homeostasis
The NMJ is a well-characterized target for small molecule effectors of neurotransmitter activity at the synaptic junction, including environmental toxins that can function as agonists and antagonists of cholinergic and GABAergic signaling. Exposure of Q35 animals to GABA, an agonist of the UNC-49 receptor, suppressed polyQ aggregation (Fig. 3C; Supplementary Fig. S5A–D), whereas exposure to lindane, an organochlorine pesticide that functions as a UNC-49 antagonist, enhanced polyQ aggregation (Fig. 3D; Supplementary Fig. S5E–H). Likewise, incubation with ACh agonists nicotine (Fig. 3F; Supplementary Fig. S5M–P) or levamisole (Fig. 3E; Supplementary Fig. S5I–L) also enhanced polyQ aggregation. We also observed similar effects of these pharmacological agents on polyQ aggregation in Q35;unc-30(rm7) animals (data not shown). The effects of the different compounds on polyQ aggregation were slightly weaker than the effects of the GABAergic mutants, and similar to what we had observed with the ACh genetic data, These results reveal that small molecule agonists and antagonists are environmental factors that affect protein homeostasis in post-synaptic muscle cells as a result of a chemical imbalance in neuronal signaling. Taken together, our data show that an imbalance in cholinergic signaling has deleterious effects on protein homeostasis whether induced by a genetic deficiency or small molecules.
Cellular hyperstimulation has global consequences on the conformation and activity of metastable proteins
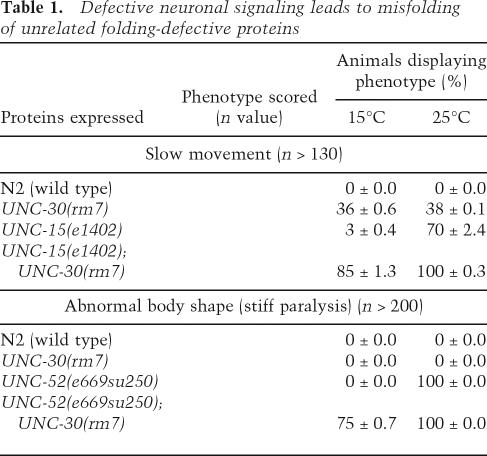
Our results suggest a model in which neuronal signaling modulates protein homeostasis. To examine whether the imbalance in neuronal signaling has global deleterious effects on protein homeostasis, we examined the phenotypes of two metastable proteins with temperature-sensitive (ts) mutations: unc-15(e1402), an allele of paramyosin, and unc-52(e669su250), a ts allele of perlecan with a role in muscle structure (Gidalevitz et al. 2006). At the restrictive temperature, the folding defect of a ts allele of unc-15(e1402) can be monitored both at a cellular level by the failure to form myofilaments and behaviorally by measuring uncoordinated movement (Gengyo-Ando and Kagawa 1991). Conversely, the folding defect caused by the unc-52(e669su250) ts mutation results in muscle dysfunction and stiff paralysis (Rogalski et al. 2001). The phenotypes of both ts strains were determined in the background of unc-30(rm7) mutants. If unc-30 causes a general disruption in protein homeostasis, we would predict that these ts phenotypes will be expressed at the permissive conditions.
For each of these ts mutations we observed a strong mutant phenotype at the restrictive condition: UNC-52(ts) animals become paralyzed except for the head region, and likewise UNC-15(ts) become uncoordinated (Table 1). At the permissive conditions, these ts phenotypes are either not detected or only weakly penetrant (Table 1; Fig. 4A,B). However, when each ts protein was expressed in the background of unc-30(rm7), we observed the ts phenotypes for both double mutants at the permissive condition (Table 1; Fig. 4A,B). A striking increase in the fraction of uncoordinated animals, from 36% to 85% was observed for unc-15(e1402);unc-30(rm7) relative to unc-30(rm7) alone (Table 1; Fig. 4A). A similar severe effect was observed for unc-52(e669su250);unc-30(rm7) in which 75% of the population displayed paralysis relative to none for control animals (Fig. 4B).
Table 1.
Defective neuronal signaling leads to misfolding of unrelated folding-defective proteins
Figure 4.
Cellular hyperstimulation exposes ts phenotypes at permissive temperatures. Age-synchronized animals were kept at 15°C up to 6 d of age, when they were analyzed for different phenotypes. (A) The strains tested for slow movement were unc-15(e1402) and unc-15(e1402);unc-30(rm7). (B) The strains unc-52(e669su250) and unc-52(e669su250);unc-30(rm7) were scored for abnormal body shape and paralysis.The wild type (N2) and unc-30(rm7) were used as controls for both assays. Graph bars represent the percentage of scored phenotype at 15°C for at least 130 animals. For both graphs, error bars indicate SEM. (C–H) Confocal projections (20×) of body wall muscle cells stained with antibody against paramyosin are shown. At the permissive temperature (15°C), the thick filaments in unc-15(e1402) animals exhibit the characteristic muscle cell structure indistinguishable from wild-type animals, whereas at the restrictive temperature disruption of muscle structure was observed in all unc-15(e1402) animals due to improper formation of thick filaments. (C) unc-15(e1402) at 15°C. (D) unc-30(rm7) at 15°C. (E) unc-15(e1402);unc-30(rm7) at 15°C. (F) unc-15(e1402) at 25°C. (G) unc-30(rm7) at 25°C. (H) unc-15(e1402);unc-30(rm7) at 25°C. All images were taken from the midsection of day 6 animals. Bar: A–F, 5 μm.
We next examined the specificity of the loss of protein homeostasis caused by the unc-30(rm7) mutation on unc-15(e1402) function by the direct examination of paramyosin in thick filament structure by immunostaining. At the permissive temperature (15°C), the thick filaments in unc-15(e1402) animals exhibit the characteristic muscle cell structure indistinguishable from wild-type animals (Fig. 4C), whereas at the restrictive temperature there is extensive disruption of muscle structure in all unc-15(e1402) animals (100%, n = 5) due to improper formation of thick filaments (Fig. 4F). At both the permissive and restrictive temperatures, unc-30(rm7) worms have a wild-type muscle structure (100%, n = 4 and 100%, n = 6) (Fig. 4D,G, respectively), whereas in double-mutant animals unc-15(e1402);unc-30(rm7) the structure of thick filaments is completely disrupted at the permissive temperature (100%, n = 7) (Fig. 4E). These results indicate that the presence of the unc-30 mutation exposes the misfolding defect of a metastable protein with a ts mutation, disrupting muscle structure (cellular level) and muscle function (behavioral level). From these results, we conclude that neuronal signaling has important consequences on protein homeostasis and interferes with the folding and activity of proteins with mild folding defects.
Discussion
Neuronal overexcitation of post-synaptic muscle cells, whether caused by an imbalance of GABAergic or cholinergic signaling, compromises protein homeostasis. This was revealed by identification of a mutation in unc-30 and the subsequent demonstration that other mutations that alter neuromuscular transmission led to misfolding of polyQ expansion proteins and other metastable proteins harboring ts mutations. These observations establish that the problem of protein folding and maintenance of protein homeostasis is not solely the result of intrinsic cellular events, but can also be influenced significantly in a non-cell-autonomous manner by perturbations in neuronal cell signaling. Our data support a model (Fig. 5) in which cellular overstimulation has severe consequences on the folding stability of proteins, and that an imbalance in neuronal signaling may be a common feature of protein conformation disorders. The non-cell-autonomous regulation of post-synaptic cellular homeostasis is likely to be of central importance not only to neurodegenerative and neuromuscular disease but may have widespread relevance in systemic pathologies associated with aging. We further propose that small molecules that alter neurotransmission, some of which correspond to widespread environmental contaminants, represent a new class of modifiers of conformational disorders.
Figure 5.
Neuronal signaling modulates the post-synaptic cell’s capacity for protein folding. We propose that an imbalance in neuronal signaling resulting in overstimulation of post-synaptic cells disrupts the cell’s folding capacity and causes misfolding and aggregation of metastable and aggregation-prone proteins. (ACh) Acetylcholine; (AChR) acetylcholine receptor; (GABA) γ-aminobutyric acid; (GABAR) GABA receptor.
Neuronal signaling as a non-cell-autonomous modulator of protein homeostasis
The ability of the cellular machinery to prevent accumulation of damaged, misfolded or aggregated proteins is influenced by the genetic background. Genome-wide genetic screens for modulators of protein aggregation have identified ∼200 genes, providing an initial understanding of the complexity of cellular protein homeostasis (Nollen et al. 2004). These results revealed how orchestration among diverse cellular processes can act to sense and restore balance to the protein folding environment in the cell (Nollen et al. 2004). The identification of neuronal signaling pathways as modulators of protein misfolding and aggregation is unexpected. Previous work has linked an ACh-regulated response to starvation and described an enhancement in protein catabolism in a subset of muscle cells that was suggested to provide the metabolic resources necessary for organismal survival (Szewczyk et al. 2000). It has also been shown that loss of both GABA and ACh signaling to the post-synaptic cells led to selective targeting of GABAA receptors for autophagy (Rowland et al. 2006). While it is well established that protein folding processes can be influenced by external stimuli—for example, heat shock or oxidative stress—the regulation of protein homeostasis has been traditionally viewed as an intrinsic process. These results reveal that protein homeostasis can be strongly influenced by extracellular communication and, in particular, neuronal activity.
We only examined body wall muscle cells as one class of post-synaptic cells, but we suggest that the neural modulatory effects on protein homeostasis may constitute a regulatory mechanism of broader significance. The ability of the cell to maintain a proper protein folding environment may constitute an important early sensor of an imbalance in presynaptic neuronal activity, before progressive disruption and irreversible cellular damage occurs. Our studies also show that neuronal excitation results in the chronic exposure of the post-synaptic cell to higher levels of intracellular Ca2+ that has a myriad of effects on cellular processes including protein homeostasis. Cell–cell communication, and the intracellular changes these processes elicit, thus emerges as an additional level of regulation and integration of different inputs in a multicellular organism with potential to have an effect on the organism’s general fitness.
While we employed ts proteins as a tool to monitor protein homeostasis, polymorphisms are abundant in the eukaryotic proteome (Goodstadt and Ponting 2001) and their penetrance have been suggested to depend on the robustness of the protein quality control mechanisms in the cell (Sangster et al. 2004; Kimchi-Sarfaty et al. 2007).
In disparate model organisms, the disruption of quality control mechanisms, by either chronic expression of aggregation-prone proteins or the loss of Hsp90 function, have been shown to cause an imbalance in cellular homeostasis, exposing otherwise silent genetic variation (Gidalevitz et al. 2006). In post-reproductive aging organisms, the exposure of silent genetic variation, due to the accumulative effects of various stresses, may contribute to disease susceptibility (Halapi and Hakonarson 2002; Mularoni et al. 2006; Kimchi-Sarfaty et al. 2007). Indeed, the majority of conformational disorders are age related and caused by the chronic expression of aggregation-prone proteins (Dobson 2003; Soto 2003). Here, we reveal that an imbalance in cell–cell communication introduces a form of chronic cell stress that can destabilize cellular homeostasis and affect disease progression.
Small molecule effectors of neuronal excitation as modulators of protein conformational disorders
Despite the prominence of protein misfolding as a causative agent of conformational disorders, another common feature of neurodegenerative diseases is the association with other environmental factors that contribute to disease onset and progression (Soto 2003). Examples include Huntington’s disease, for which patients with the same polyQ length can exhibit variation in age of disease onset (Wexler et al. 2004), Parkinson’s disease, in which post-industrial environmental exposures such as insecticides have been correlated with disease onset (Benmoyal-Segal and Soreq 2006), and ALS, in which the majority of the cases are sporadic (Bruijn et al. 2004).
Our results reveal that small molecule agonists and antagonists that alter the balance of ACh and GABA at the NMJ cause an imbalance in protein homeostasis in post-synaptic cells, suggesting an important new role for commonly ingested chemicals (nicotine) and other widespread environmental contaminants (lindane) with neurostimulatory properties in protein misfolding diseases. Organochlorine compounds and other related toxicological agents have been implicated in diverse human diseases (Bruijn et al. 2004; Zawia and Basha 2005; Benmoyal-Segal and Soreq 2006). The chronic exposure to these compounds could, over time, lead to a progressive imbalance in protein homeostasis that exacerbates the proper folding and clearance of conformationally challenged proteins. Consequently such environmental toxicants could contribute significantly to the onset of sporadic diseases such as ALS, for which an imbalance in GABAergic synaptic transmission seems to be pivotal for differential vulnerability of particular sets of cerebrospinal axis motor neurons (Lorenzo et al. 2006). Whether such compounds are directly causal or not, our data would support their roles to imbalance homeostasis as potential modifiers of disease.
Materials and methods
Strains
Nematodes were handled using standard methods (Brenner 1974) and experiments were performed at 20°C, unless otherwise indicated. Strains and alleles used in this study were as follows: Bristol strain N2 (wild type), unc-30(e191), unc-30(e2327), unc-25(e156), unc-47(gk192), unc-49(e407), Punc-47∷ gfp, rmIs132[Punc-54∷q35∷yfp], rmIs133[Punc-54∷q40∷yfp], ace-1(p1000), ace-2(g72), egl-19(ad692), unc-15(e1402), and unc-52(e669su250). The unc-30(e2327) strain was kindly provided by the Horvitz laboratory; all other strains are available from the Caenorhabditis elegans Genetic Center. Strains containing Q35 and Q40 stretches were described previously (Morley et al. 2002; Nollen et al. 2004). Strains generated in this work are listed in the Supplemental Material.
Mutagenesis, mapping, and molecular analysis of unc-30(rm7)
Mutagenesis with ethyl methanesulfonate (EMS) was performed by standard methods (Brenner 1974). unc-30(rm7) chromosome mapping was performed by SNPs (Wicks et al. 2001). UNC-30 function rescue was obtained by coinjecting the cosmid B0564 (40 μg/μL) and a cotransformational marker Pmyo-2∷gfp (100 ng/μl) (gift from A. Fire, Stanford University, Stanford, CA) in unc-30(rm7) animals. Transgenic lines were identified based on the presence of the fluorescent marker and transgenic animals were analyzed for aggregation and uncoordination phenotypes [detailed identification of unc-30(rm7) is described in the Supplemental Material].
Fluorescence microscopy and fluorescence recovery after photobleaching
For in vivo imaging, animals were mounted on a 2% agar pad on a glass slide and immobilized in 1 mM levamisole (Sigma). Fluorescence micrographs were taken with a Zeiss Axioplan 2/LSM 510 Meta confocal microscope. Immobilized animals were subjected to FRAP analysis as described (Nollen et al. 2004) with the following modification: An area of 3.6 μm2 was bleached for 6.4 sec (25 iterations at 100% laser power), after which an image was collected every 30 sec (at 0.1% laser power). Relative fluorescence intensity (RFI) was determined by using the equation RFI = (Net/N1t)/(Ne0/N10). Net is the average intensity of the bleached area at a given time point, and N1t is the average intensity of an adjacent nonbleached area at the corresponding time point, allowing to control for general photobleaching and background fluorescence. Ne0 and N10 are the average intensity before photobleaching of the bleached and nonbleached control area, respectively.
Aggregate quantification
Animals were age-synchronized by bleaching with NaOCl and left overnight to hatch on either unseeded NGM plates or in S-medium for the drug assays. For the Q40 or Q35 signaling-defective backgrounds, single L1-stage animals were picked and put onto individual seeded plates for aggregate counting and the same animal was scored for aggregation during 6 or 10 d, respectively. A minimum of 21 animals were scored for each strain and for each time point analyzed. For the drug assays, a population study was performed. At each time point animals were transferred from 96-well microtiter liquid cultures onto plates and the total number of aggregates was scored for each individual. For RNA interference (RNAi) assays, synchronized Q35 L1 animals were transferred onto individual NGM plates seeded with Escherichia coli containing the appropriate target sequence or vector alone, L4440. Aggregates were counted for each animal over time. Staged animals were viewed at 100× magnification with a stereomicroscope equipped for epifluorescence, and the number of polyQ aggregates was determined. Aggregates were defined as discrete, bright structures, with boundaries distinguishable from surrounding fluorescence. The number of aggregates for each animal in each day was counted at least twice and the values were averaged. For the drug assays, the quantification was performed blind. Data were collected from different experiments and compiled to calculate daily aggregate number averages. Day 1 is defined as 24 h following NaOCl age synchronization and animals are said to be 1 d old.
Assay for ts phenotypes
For the slow movement assay, age-synchronized L1 nematodes were grown at 15°C and scored 5 d later. Per assay, eight to 12 animals were placed in a freshly seeded plate (equilibrated to 20°C) in the center of a 1-cm ring. Animals remaining in the 1-cm circle after 5 min were considered to possess a slow movement phenotype (Gidalevitz et al. 2006). As a control experiment, L1 nematodes were grown at 15°C until L3–L4 stage (to avoid embryonic and developmental phenotypes), transferred to 25°C, and scored 1 d later for the same phenotype. Experiments were repeated until the n value was at least 130. To score for abnormal body shape (or stiff paralysis), age-synchronized L1 nematodes were grown for 6 d at 15°C. Partially paralyzed animals with moving heads and stick-like bodies were scored for abnormal body shape. The controls were grown at 15°C until L3–L4 stage, transferred to 25°C, and scored 2 d later.
Immunofluorescence analysis
Six-day-old nematodes were collected using M9 buffer and fixed for 2 h in 40 mg/mL formaldehyde/phosphate buffer (pH 7.2) at 4°C. After fixation, animals were washed in PBS (pH 7.4) and incubated in 2% Tween20 for 30 min at room temperature. For permeabilization, animals were washed in PBS (pH 7.4), resuspended in 5% 2-Mercaptoethanol/Tris (pH 6.8) (with 1% Triton X-100), and incubated overnight at 37°C. Samples were then treated with 2.5 U/μL Collagenase/Tris (pH 7.4) (type IV, Sigma-Aldrich #C5138) for ∼30 min at 37°C, using a shaker at 250 rpm. This reaction was stopped by transferring the samples to ice. Fixed-permeabilized samples were blocked with AbA solution (10 mg/mL BSA in PBS) followed by antibody staining. Samples were incubated overnight with primary antibody against paramyosin (1:200 dilution, Developmental Studies Hybridoma Bank mouse IgG1, gift from H. Epstein, University of Texas, Galveston, TX) at room temperature. Incubation with secondary antibody was performed for 2 h using anti-mouse IgG-FITC conjugate (1:200 dilution; Sigma #F0257). Nematodes were mounted in 90% glycerol/1 mg/mL DABCO (diaza-bicyclo-octane) anti-fading solution. Samples were examined by immunofluorescence microscopy using a Zeiss ConfoCor3/510 Meta confocal microscope (Ruvkun and Giusto 1989; Finney and Ruvkun 1990).
Drug assays
Drug assays were performed in 96-well plates in liquid culture (Voisine et al. 2007). Each well contained a final volume of 60 μL comprising 10–15 L1 animals, drugs at the appropriate concentration, and HB101 bacteria to a final OD of 0.8 in the microtiter plate. Animals and bacteria were resuspended in S-medium supplemented with streptomycin, penicillin, and nystatin (Sigma). C. elegans were grown with continuous shaking at 200 rpm at 20°C (New Brunswick Scientific Incubation shaker). Compound preparation: Stock solutions of 500 mM GABA (Sigma) and 800 mM levamisole (Sigma) were prepared in water; stock solutions of 300 mM nicotine (Sigma) and 170 mM lindane (Sigma) were prepared in ethanol.
Acknowledgments
We thank members of the Morimoto Laboratory and Dr. Robert Holmgren for discussions and comments on the manuscript, and J. Merchant, C. Clendaniel, and A. Ben-Zvi for assistance in these studies. We also thank J. West for advice in statistical analysis. We are grateful to T. O’Halloran’s laboratory for the use of a Zeiss confocal microscope and the Biological Imaging Facility at Northwestern University. We acknowledge T. Stiernagle and the Caenorhabditis elegans Genetics Center—supported by a grant from the National Institutes of General Medical Sciences—and the International C. elegans Gene Knockout Consortium for providing C. elegans strains. We also thank A. Fire and R. Horvitz for providing strains and plasmids. The monoclonal antibody anti-paramyosin developed by H. Epstein was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA. S.M.G. is supported by Ph.D. fellowship PRAXIS XXI BD/21451/99 FCT/FEDER (Portugal/EU); M.D.A. is supported by POCTI/BIA-BCM/56609/2004 grant and CIGMH FCT/FEDER (Portugal/EU); M.O.C. is supported by a post-doctoral fellowship from the American Parkinson Disease Association; and M.C.S. is supported by Ph.D. fellowship SFRH/BD/28461/2006 FCT/FEDER (Portugal/EU). These studies were supported by grants to R.I.M. from the NIH (NIGMS and NIA), the Huntington’s Disease Society of America Coalition for the Cure, and the Daniel F. and Ada L. Rice Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1575307
References
- Amaral M.D. Therapy through chaperones: Sense or antisense? Cystic fibrosis as a model disease. J. Inherit. Metab. Dis. 2006;29:477–487. doi: 10.1007/s10545-006-0251-x. [DOI] [PubMed] [Google Scholar]
- Benmoyal-Segal L., Soreq H., Soreq H. Gene–environment interactions in sporadic Parkinson’s disease. J. Neurochem. 2006;97:1740–1755. doi: 10.1111/j.1471-4159.2006.03937.x. [DOI] [PubMed] [Google Scholar]
- Boillee S., Vande Velde C., Cleveland D.W., Vande Velde C., Cleveland D.W., Cleveland D.W. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull H.R., Moore F.E., Tang S.J., Morimoto R.I., Moore F.E., Tang S.J., Morimoto R.I., Tang S.J., Morimoto R.I., Morimoto R.I. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J. Neurosci. 2006;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull H.R., Morley J.F., Morimoto R.I., Morley J.F., Morimoto R.I., Morimoto R.I. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv. Exp. Med. Biol. 2007;594:167–189. doi: 10.1007/978-0-387-39975-1_15. [DOI] [PubMed] [Google Scholar]
- Bruijn L.I., Miller T.M., Cleveland D.W., Miller T.M., Cleveland D.W., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Chen S., Ferrone F.A., Wetzel R., Ferrone F.A., Wetzel R., Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen F.E., Kelly J.W., Kelly J.W. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- Combes D., Fedon Y., Toutant J.P., Arpagaus M., Fedon Y., Toutant J.P., Arpagaus M., Toutant J.P., Arpagaus M., Arpagaus M. Multiple ace genes encoding acetylcholinesterases of Caenorhabditis elegans have distinct tissue expression. Eur. J. Neurosci. 2003;18:497–512. doi: 10.1046/j.1460-9568.2003.02749.x. [DOI] [PubMed] [Google Scholar]
- de la Cruz I.P., Levin J.Z., Cummins C., Anderson P., Horvitz H.R., Levin J.Z., Cummins C., Anderson P., Horvitz H.R., Cummins C., Anderson P., Horvitz H.R., Anderson P., Horvitz H.R., Horvitz H.R. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J. Neurosci. 2003;23:9133–9145. doi: 10.1523/JNEUROSCI.23-27-09133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Drysdale M.J., Brough P.A., Massey A., Jensen M.R., Schoepfer J., Brough P.A., Massey A., Jensen M.R., Schoepfer J., Massey A., Jensen M.R., Schoepfer J., Jensen M.R., Schoepfer J., Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr. Opin. Drug Discov. Devel. 2006;9:483–495. [PubMed] [Google Scholar]
- Faber P.W., Alter J.R., MacDonald M.E., Hart A.C., Alter J.R., MacDonald M.E., Hart A.C., MacDonald M.E., Hart A.C., Hart A.C. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K., Kagawa H., Kagawa H. Single charge change on the helical surface of the paramyosin rod dramatically disrupts thick filament assembly in Caenorhabditis elegans. J. Mol. Biol. 1991;219:429–441. doi: 10.1016/0022-2836(91)90184-8. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I., Ho K.H., Brignull H.R., Morimoto R.I., Brignull H.R., Morimoto R.I., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Goodstadt L., Ponting C.P., Ponting C.P. Sequence variation and disease in the wake of the draft human genome. Hum. Mol. Genet. 2001;10:2209–2214. doi: 10.1093/hmg/10.20.2209. [DOI] [PubMed] [Google Scholar]
- Gregersen N. Protein misfolding disorders: Pathogenesis and intervention. J. Inherit. Metab. Dis. 2006;29:456–470. doi: 10.1007/s10545-006-0301-4. [DOI] [PubMed] [Google Scholar]
- Gusella J.F., MacDonald M.E., MacDonald M.E. Molecular genetics: Unmasking polyglutamine triggers in neurodegenerative disease. Nat. Rev. Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- Halapi E., Hakonarson H., Hakonarson H. Advances in the development of genetic markers for the diagnosis of disease and drug response. Expert Rev. Mol. Diagn. 2002;2:411–421. doi: 10.1586/14737159.2.5.411. [DOI] [PubMed] [Google Scholar]
- Hsu A.L., Murphy C.T., Kenyon C., Murphy C.T., Kenyon C., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jin Y., Hoskins R., Horvitz H.R., Hoskins R., Horvitz H.R., Horvitz H.R. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- Jin Y., Jorgensen E., Hartwieg E., Horvitz H.R., Jorgensen E., Hartwieg E., Horvitz H.R., Hartwieg E., Horvitz H.R., Horvitz H.R. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J. Neurosci. 1999;19:539–548. doi: 10.1523/JNEUROSCI.19-02-00539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jospin M., Jacquemond V., Mariol M.C., Segalat L., Allard B., Jacquemond V., Mariol M.C., Segalat L., Allard B., Mariol M.C., Segalat L., Allard B., Segalat L., Allard B., Allard B. The L-type voltage-dependent Ca2+ channel EGL-19 controls body wall muscle function in Caenorhabditis elegans. J. Cell Biol. 2002;159:337–348. doi: 10.1083/jcb.200203055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Ambudkar S.V., Gottesman M.M., Gottesman M.M. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Lee R.Y., Lobel L., Hengartner M., Horvitz H.R., Avery L., Lobel L., Hengartner M., Horvitz H.R., Avery L., Hengartner M., Horvitz H.R., Avery L., Horvitz H.R., Avery L., Avery L. Mutations in the α1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. EMBO J. 1997;16:6066–6076. doi: 10.1093/emboj/16.20.6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo L.E., Barbe A., Portalier P., Fritschy J.M., Bras H., Barbe A., Portalier P., Fritschy J.M., Bras H., Portalier P., Fritschy J.M., Bras H., Fritschy J.M., Bras H., Bras H. Differential expression of GABAA and glycine receptors in ALS-resistant vs. ALS-vulnerable motoneurons: Possible implications for selective vulnerability of motoneurons. Eur. J. Neurosci. 2006;23:3161–3170. doi: 10.1111/j.1460-9568.2006.04863.x. [DOI] [PubMed] [Google Scholar]
- McIntire S.L., Reimer R.J., Schuske K., Edwards R.H., Jorgensen E.M., Reimer R.J., Schuske K., Edwards R.H., Jorgensen E.M., Schuske K., Edwards R.H., Jorgensen E.M., Edwards R.H., Jorgensen E.M., Jorgensen E.M. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- Morley J.F., Brignull H.R., Weyers J.J., Morimoto R.I., Brignull H.R., Weyers J.J., Morimoto R.I., Weyers J.J., Morimoto R.I., Morimoto R.I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L., Guigo R., Alba M.M., Guigo R., Alba M.M., Alba M.M. Mutation patterns of amino acid tandem repeats in the human proteome. Genome Biol. 2006;7:R33. doi: 10.1186/gb-2006-7-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen E.A., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Chavez A., Morimoto R.I., Plasterk R.H., Morimoto R.I., Plasterk R.H., Plasterk R.H. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.A., Connolly J.B., Wellington C., Hayden M., Dausset J., Neri C., Connolly J.B., Wellington C., Hayden M., Dausset J., Neri C., Wellington C., Hayden M., Dausset J., Neri C., Hayden M., Dausset J., Neri C., Dausset J., Neri C., Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc. Natl. Acad. Sci. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T.A., Lindquist S., Sangster T.A., Lindquist S., Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Richmond J.E., Jorgensen E.M., Jorgensen E.M. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat. Neurosci. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski T.M., Mullen G.P., Bush J.A., Gilchrist E.J., Moerman D.G., Mullen G.P., Bush J.A., Gilchrist E.J., Moerman D.G., Bush J.A., Gilchrist E.J., Moerman D.G., Gilchrist E.J., Moerman D.G., Moerman D.G. UNC-52/perlecan isoform diversity and function in Caenorhabditis elegans. Biochem. Soc. Trans. 2001;29:171–176. doi: 10.1042/0300-5127:0290171. [DOI] [PubMed] [Google Scholar]
- Rowland A.M., Richmond J.E., Olsen J.G., Hall D.H., Bamber B.A., Richmond J.E., Olsen J.G., Hall D.H., Bamber B.A., Olsen J.G., Hall D.H., Bamber B.A., Hall D.H., Bamber B.A., Bamber B.A. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J. Neurosci. 2006;26:1711–1720. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusmini P., Sau D., Crippa V., Palazzolo I., Simonini F., Onesto E., Martini L., Poletti A., Sau D., Crippa V., Palazzolo I., Simonini F., Onesto E., Martini L., Poletti A., Crippa V., Palazzolo I., Simonini F., Onesto E., Martini L., Poletti A., Palazzolo I., Simonini F., Onesto E., Martini L., Poletti A., Simonini F., Onesto E., Martini L., Poletti A., Onesto E., Martini L., Poletti A., Martini L., Poletti A., Poletti A. Aggregation and proteasome: The case of elongated polyglutamine aggregation in spinal and bulbar muscular atrophy. Neurobiol. Aging. 2007;28:1099–1111. doi: 10.1016/j.neurobiolaging.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Rutherford S.L., Lindquist S., Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Ruvkun G., Giusto J., Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Sangster T.A., Lindquist S., Queitsch C., Lindquist S., Queitsch C., Queitsch C. Under cover: Causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- Satyal S.H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I., Kitagawa K., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I., Lindquist S., Kramer J.M., Morimoto R.I., Kramer J.M., Morimoto R.I., Morimoto R.I. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Stefani M. Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Szewczyk N.J., Hartman J.J., Barmada S.J., Jacobson L.A., Hartman J.J., Barmada S.J., Jacobson L.A., Barmada S.J., Jacobson L.A., Jacobson L.A. Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J. Cell. Sci. 2000;113:2003–2010. doi: 10.1242/jcs.113.11.2003. [DOI] [PubMed] [Google Scholar]
- Voisine C., Varma H., Walker N., Bates E.A., Stockwell B.R., Hart A.C., Varma H., Walker N., Bates E.A., Stockwell B.R., Hart A.C., Walker N., Bates E.A., Stockwell B.R., Hart A.C., Bates E.A., Stockwell B.R., Hart A.C., Stockwell B.R., Hart A.C., Hart A.C. Identification of potential therapeutic drugs for Huntington’s disease using Caenorhabditis elegans. PLoS ONE. 2007;2:e504. doi: 10.1371/journal.pone.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J.M., Paulson H.L., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M., Paulson H.L., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M., Fischbeck K.H., Pittman R.N., Bonini N.M., Pittman R.N., Bonini N.M., Bonini N.M. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- Wexler N.S., Lorimer J., Porter J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Lorimer J., Porter J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Porter J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Marder K., Penchaszadeh G., Roberts S.A., Gayan J., Penchaszadeh G., Roberts S.A., Gayan J., Roberts S.A., Gayan J., Gayan J., et al. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.G., Albertson D.G., Anness M.A., Albertson D.G., Anness M.A., Anness M.A. Connectivity changes in a class of motoneurone during the development of a nematode. Nature. 1978;271:764–766. doi: 10.1038/271764a0. [DOI] [PubMed] [Google Scholar]
- Wicks S.R., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Yeh R.T., Gish W.R., Waterston R.H., Plasterk R.H., Gish W.R., Waterston R.H., Plasterk R.H., Waterston R.H., Plasterk R.H., Plasterk R.H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zawia N.H., Basha M.R., Basha M.R. Environmental risk factors and the developmental basis for Alzheimer’s disease. Rev. Neurosci. 2005;16:325–337. doi: 10.1515/revneuro.2005.16.4.325. [DOI] [PubMed] [Google Scholar]