Abstract
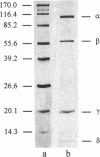
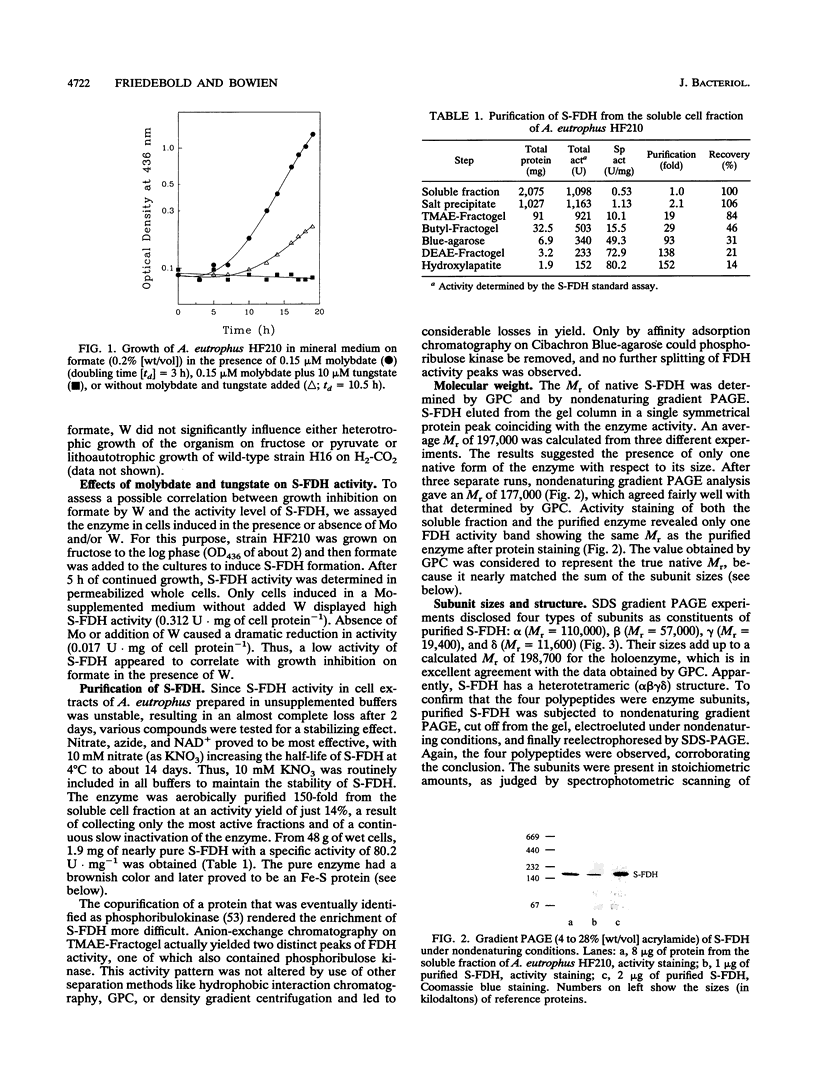
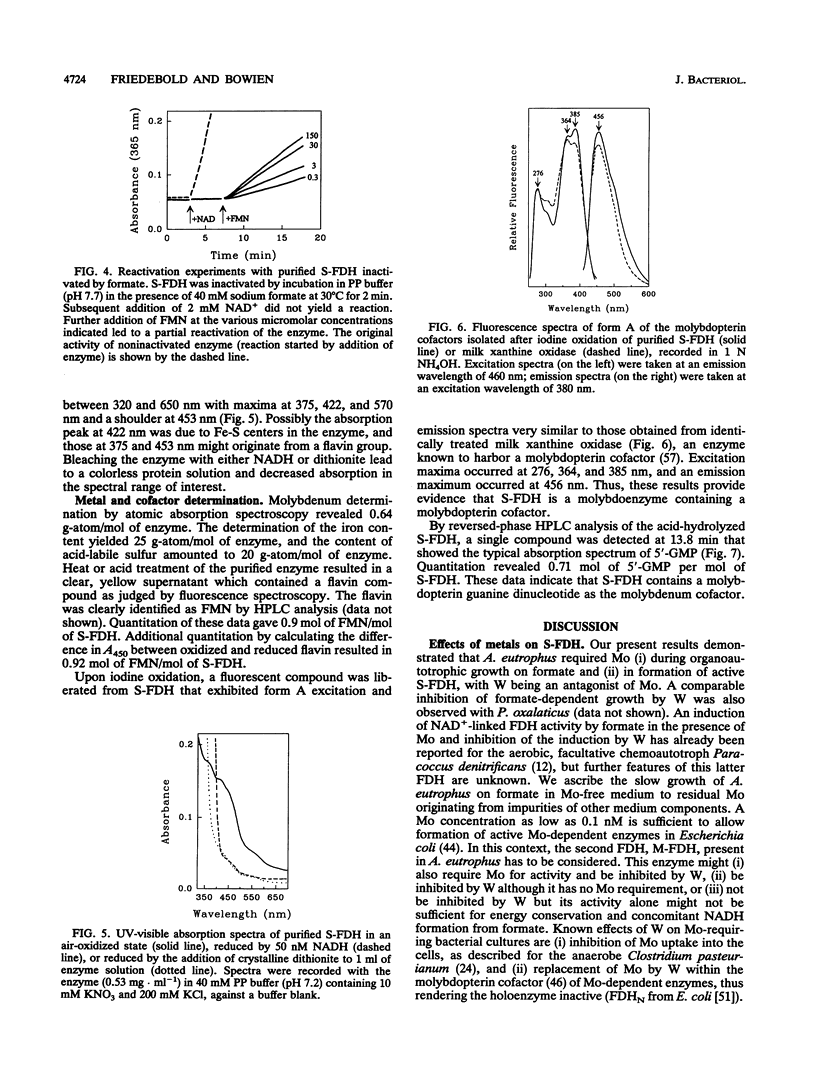
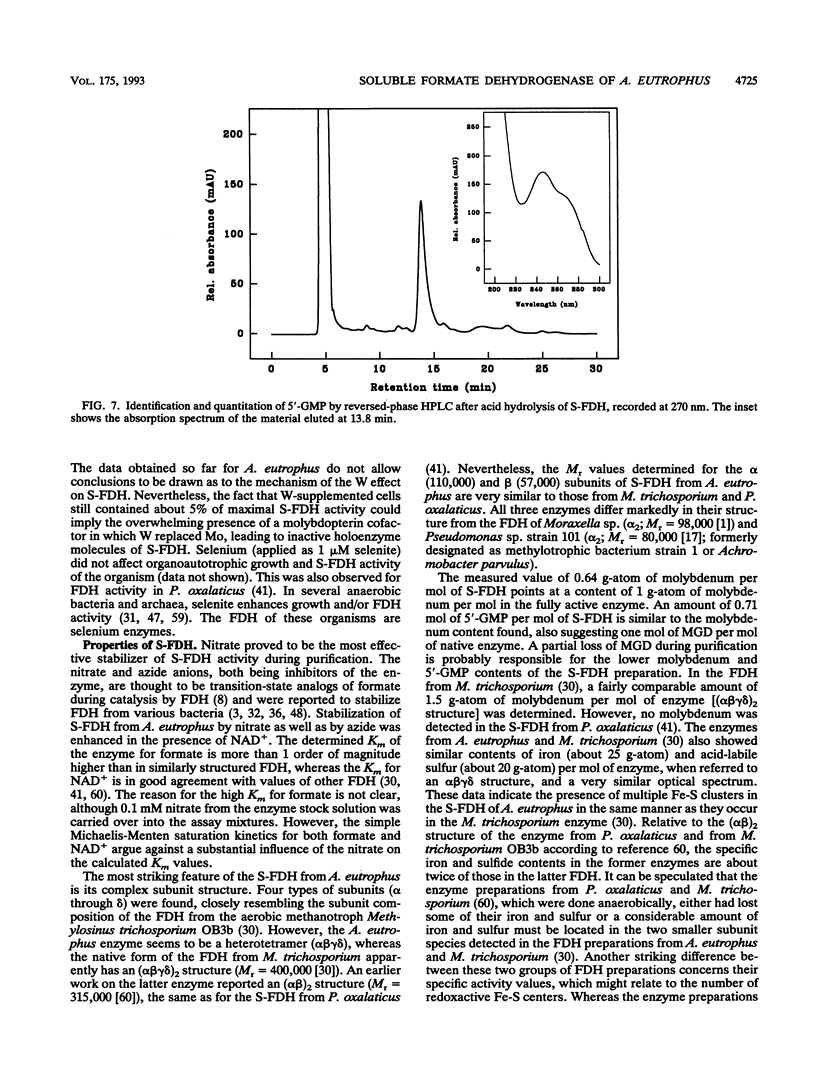
Organoautotrophic growth of Alcaligenes eutrophus on formate was dependent on the presence of molybdate in the medium. Supplementation of the medium with tungstate lead to growth cessation. Corresponding effects of these anions were observed for the activity of the soluble, NAD(+)-linked formate dehydrogenase (S-FDH; EC 1.2.1.2) of the organism. Lack of molybdate or presence of tungstate resulted in an almost complete loss of S-FDH activity. S-FDH was purified to near homogeneity in the presence of nitrate as a stabilizing agent. The native enzyme exhibited an M(r) of 197,000 and a heterotetrameric quaternary structure with nonidentical subunits of M(r) 110,000 (alpha), 57,000 (beta), 19,400 (gamma), and 11,600 (delta). It contained 0.64 g-atom of molybdenum, 25 g-atom of nonheme iron, 20 g-atom of acid-labile sulfur, and 0.9 mol of flavin mononucleotide per mol. The fluorescence spectrum of iodine-oxidized S-FDH was nearly identical to the form A spectrum of milk xanthine oxidase, proving the presence of a pterin cofactor. The molybdenum-complexing cofactor was identified as molybdopterin guanine dinucleotide in an amount of 0.71 mol/mol of S-FDH. Apparent Km values of 3.3 mM for formate and 0.09 mM for NAD+ were determined. The enzyme coupled the oxidation of formate to a number of artificial electron acceptors and was strongly inactivated by formate in the absence of NAD+. It was inhibited by cyanide, azide, nitrate, and Hg2+ ions. Thus, the enzyme belongs to a new group of complex molybdo-flavo Fe-S FDH that so far has been detected in only one other aerobic bacterium.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asano Y., Sekigawa T., Inukai H., Nakazawa A. Purification and properties of formate dehydrogenase from Moraxella sp. strain C-1. J Bacteriol. 1988 Jul;170(7):3189–3193. doi: 10.1128/jb.170.7.3189-3193.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilova T. V., Egorova O. A., Ioanesyan L. S., Egorov A. M. Biosynthesis, isolation and properties of NAD-dependent formate dehydrogenase from the yeast Candida methylica. Eur J Biochem. 1985 Nov 4;152(3):657–662. doi: 10.1111/j.1432-1033.1985.tb09245.x. [DOI] [PubMed] [Google Scholar]
- Axley M. J., Grahame D. A., Stadtman T. C. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem. 1990 Oct 25;265(30):18213–18218. [PubMed] [Google Scholar]
- Bauder R., Tshisuaka B., Lingens F. Microbial metabolism of quinoline and related compounds. VII. Quinoline oxidoreductase from Pseudomonas putida: a molybdenum-containing enzyme. Biol Chem Hoppe Seyler. 1990 Dec;371(12):1137–1144. doi: 10.1515/bchm3.1990.371.2.1137. [DOI] [PubMed] [Google Scholar]
- Beinert H. Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 1978;54:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983 Jun;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Blanchard J. S., Cleland W. W. Kinetic and chemical mechanisms of yeast formate dehydrogenase. Biochemistry. 1980 Jul 22;19(15):3543–3550. doi: 10.1021/bi00556a020. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burke K. A., Calder K., Lascelles J. Effects of molybdenum and tungsten on induction of nitrate reductase and formate dehydrogenase in wild type and mutant Paracoccus denitrificans. Arch Microbiol. 1980 Jun;126(2):155–159. doi: 10.1007/BF00511221. [DOI] [PubMed] [Google Scholar]
- Chandra T. S., Shethna Y. I. Oxalate and formate in Alcaligenes and Pseudomonas species. Antonie Van Leeuwenhoek. 1975;41(4):465–477. doi: 10.1007/BF02565090. [DOI] [PubMed] [Google Scholar]
- Deyhle R. R., Barton L. L. Nicotinamide adenine dinucleotide -- independent formate dehydrogenase in Mycobacterium phlei. Can J Microbiol. 1977 Feb;23(2):125–130. doi: 10.1139/m77-018. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen L., Knight M., Harder W. Metabolic regulation in Pseudomonas oxalaticus OX1. Autotrophic and heterotrophic growth on mixed substrates. Arch Microbiol. 1978 Jan 23;116(1):77–83. doi: 10.1007/BF00408736. [DOI] [PubMed] [Google Scholar]
- Egorov A. M., Avilova T. V., Dikov M. M., Popov V. O., Rodionov Y. V., Berezin I. V. NAD-dependent formate dehydrogenase from methylotrophic bacterium, strain 1. Purification and characterization. Eur J Biochem. 1979 Sep;99(3):569–576. doi: 10.1111/j.1432-1033.1979.tb13289.x. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. Formate dehydrogenase from Escherichia coli. Methods Enzymol. 1982;89(Pt 500):537–543. doi: 10.1016/s0076-6879(82)89093-9. [DOI] [PubMed] [Google Scholar]
- Ferry J. G. Formate dehydrogenase. FEMS Microbiol Rev. 1990 Dec;7(3-4):377–382. doi: 10.1111/j.1574-6968.1990.tb04940.x. [DOI] [PubMed] [Google Scholar]
- Hou C. T., Patel R. N., Laskin A. I., Barnabe N. NAD-linked formate dehydrogenase from methanol-grown Pichia pastoris NRRL-Y-7556. Arch Biochem Biophys. 1982 Jun;216(1):296–305. doi: 10.1016/0003-9861(82)90214-4. [DOI] [PubMed] [Google Scholar]
- Izumi Y., Kanzaki H., Morita S., Futazuka H., Yamada H. Characterization of crystalline formate dehydrogenase from Candida methanolica. Eur J Biochem. 1989 Jun 15;182(2):333–341. doi: 10.1111/j.1432-1033.1989.tb14835.x. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Bastian N. R., Schauer N. L., Ferry J. G., Rajagopalan K. V. Identification of molybdopterin guanine dinucleotide in formate dehydrogenase from Methanobacterium formicicum. FEMS Microbiol Lett. 1991 Jan 15;61(2-3):213–216. doi: 10.1016/0378-1097(91)90554-n. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984 May 10;259(9):5414–5422. [PubMed] [Google Scholar]
- Johnson J. L., Indermaur L. W., Rajagopalan K. V. Molybdenum cofactor biosynthesis in Escherichia coli. Requirement of the chlB gene product for the formation of molybdopterin guanine dinucleotide. J Biol Chem. 1991 Jul 5;266(19):12140–12145. [PubMed] [Google Scholar]
- Jollie D. R., Lipscomb J. D. Formate dehydrogenase from Methylosinus trichosporium OB3b. Purification and spectroscopic characterization of the cofactors. J Biol Chem. 1991 Nov 15;266(32):21853–21863. [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Karzanov V. V., Bogatsky YuA, Tishkov V. I., Egorov A. M. Evidence for the presence of a new NAD+-dependent formate dehydrogenase in Pseudomonas sp. 101 cells grown on a molybdenum-containing medium. FEMS Microbiol Lett. 1989 Jul 15;51(1):197–200. doi: 10.1016/0378-1097(89)90508-9. [DOI] [PubMed] [Google Scholar]
- Karzanov V. V., Correa C. M., Bogatsky Y. G., Netrusov A. I. Alternative NAD(+)-dependent formate dehydrogenases in the facultative methylotroph Mycobacterium vaccae 10. FEMS Microbiol Lett. 1991 Jun 1;65(1):95–99. doi: 10.1016/0378-1097(91)90478-s. [DOI] [PubMed] [Google Scholar]
- Kortlüke C., Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger A., Winkler E., Innerhofer A., Hackenberg H., Schägger H. The formate dehydrogenase involved in electron transport from formate to fumarate in Vibrio succinogenes. Eur J Biochem. 1979 Mar;94(2):465–475. doi: 10.1111/j.1432-1033.1979.tb12914.x. [DOI] [PubMed] [Google Scholar]
- Krüger B., Meyer O. The pterin (bactopterin) of carbon monoxide dehydrogenase from Pseudomonas carboxydoflava. Eur J Biochem. 1986 May 15;157(1):121–128. doi: 10.1111/j.1432-1033.1986.tb09647.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu C. L., Mortenson L. E. Formate dehydrogenase of Clostridium pasteurianum. J Bacteriol. 1984 Jul;159(1):375–380. doi: 10.1128/jb.159.1.375-380.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Willnow P., Ruschig U., Höpner T. Formate dehydrogenase from Pseudomonas oxalaticus. Eur J Biochem. 1978 Feb;83(2):485–498. doi: 10.1111/j.1432-1033.1978.tb12115.x. [DOI] [PubMed] [Google Scholar]
- PINSENT J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J. 1954 May;57(1):10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock D., Boulter D. Kinetic studies of formate dehydrogenase. Biochem J. 1970 Dec;120(4):763–769. doi: 10.1042/bj1200763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poels P. A., Groen B. W., Duine J. A. NAD(P)+-independent aldehyde dehydrogenase from Pseudomonas testosteroni. A novel type of molybdenum-containing hydroxylase. Eur J Biochem. 1987 Aug 3;166(3):575–579. doi: 10.1111/j.1432-1033.1987.tb13552.x. [DOI] [PubMed] [Google Scholar]
- Rajagopalan K. V., Johnson J. L. The pterin molybdenum cofactors. J Biol Chem. 1992 May 25;267(15):10199–10202. [PubMed] [Google Scholar]
- SCHLEGEL H. G., KALTWASSER H., GOTTSCHALK G. [A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies]. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- Sawers G., Heider J., Zehelein E., Böck A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J Bacteriol. 1991 Aug;173(16):4983–4993. doi: 10.1128/jb.173.16.4983-4993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J Bacteriol. 1986 Feb;165(2):405–411. doi: 10.1128/jb.165.2.405-411.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlensog V., Böck A. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol. 1990 Aug;4(8):1319–1327. doi: 10.1111/j.1365-2958.1990.tb00711.x. [DOI] [PubMed] [Google Scholar]
- Scott R. H., DeMoss J. A. Formation of the formate-nitrate electron transport pathway from inactive components in Escherichia coli. J Bacteriol. 1976 Apr;126(1):478–486. doi: 10.1128/jb.126.1.478-486.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V. K., Ugalde R. A., Imperial J., Brill W. J. Molybdenum in nitrogenase. Annu Rev Biochem. 1984;53:231–257. doi: 10.1146/annurev.bi.53.070184.001311. [DOI] [PubMed] [Google Scholar]
- Siebert K., Schobert P., Bowien B. Purification, some catalytic and molecular properties of phosphoribulokinase from Alcaligenes eutrophus. Biochim Biophys Acta. 1981 Mar 13;658(1):35–44. doi: 10.1016/0005-2744(81)90247-3. [DOI] [PubMed] [Google Scholar]
- Uotila L., Koivusalo M. Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys. 1979 Aug;196(1):33–45. doi: 10.1016/0003-9861(79)90548-4. [DOI] [PubMed] [Google Scholar]
- Wootton J. C., Nicolson R. E., Cock J. M., Walters D. E., Burke J. F., Doyle W. A., Bray R. C. Enzymes depending on the pterin molybdenum cofactor: sequence families, spectroscopic properties of molybdenum and possible cofactor-binding domains. Biochim Biophys Acta. 1991 Mar 29;1057(2):157–185. doi: 10.1016/s0005-2728(05)80100-8. [DOI] [PubMed] [Google Scholar]
- Yagi T. Purification and properties of cytochrome c-553, an electron acceptor for formate dehydrogenase of Desulfovibrio vulgaris, Miyazaki. Biochim Biophys Acta. 1979 Oct 10;548(1):96–105. doi: 10.1016/0005-2728(79)90190-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto I., Saiki T., Liu S. M., Ljungdahl L. G. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J Biol Chem. 1983 Feb 10;258(3):1826–1832. [PubMed] [Google Scholar]
- Yoch D. C., Chen Y. P., Hardin M. G. Formate dehydrogenase from the methane oxidizer Methylosinus trichosporium OB3b. J Bacteriol. 1990 Aug;172(8):4456–4463. doi: 10.1128/jb.172.8.4456-4463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]