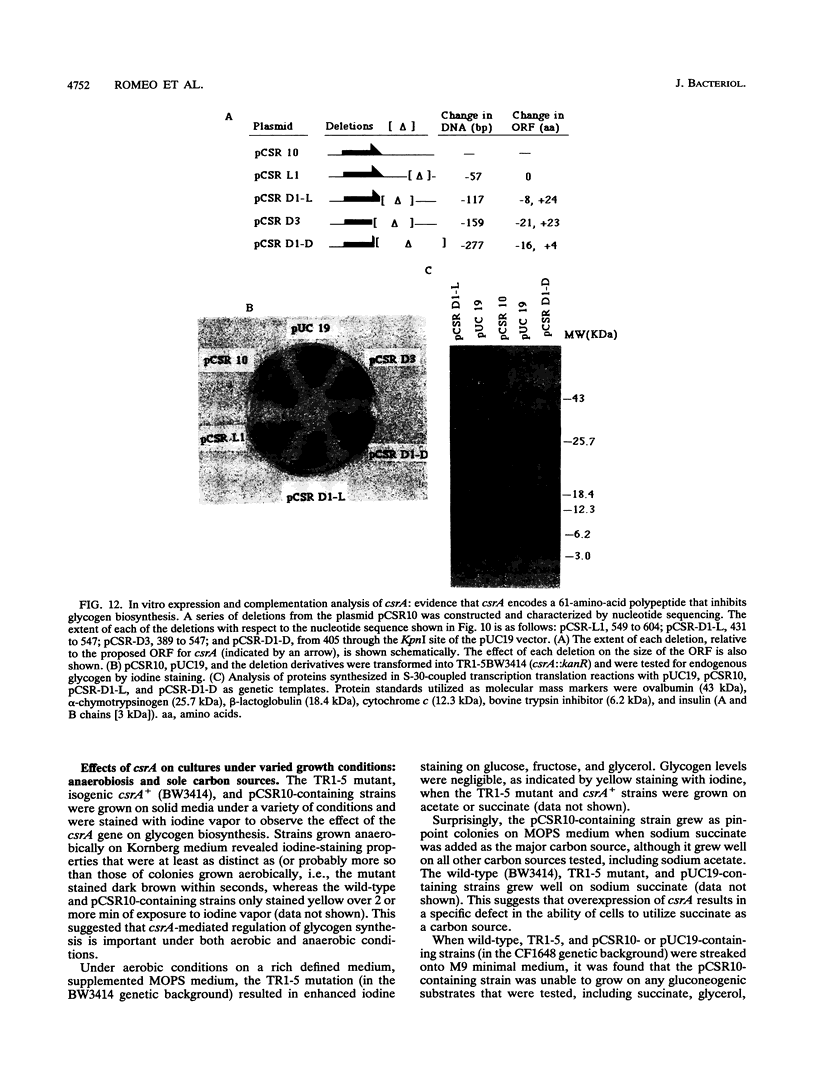
Abstract
Current evidence suggests that a few global regulatory factors mediate many of the extensive changes in gene expression that occur as Escherichia coli enters the stationary phase. One of the metabolic pathways that is transcriptionally activated in the stationary phase is the pathway for biosynthesis of glycogen. To identify factors that regulate glycogen biosynthesis in trans, a collection of transposon mutants was generated and screened for mutations which independently increase or decrease glycogen levels and the expression of a plasmid-encoded glgC'-lacZ fusion. The glycogen excess mutation TR1-5 was found to be pleiotropic. It led to increased expression of the genes glgC (ADPglucose pyrophosphorylase) and glgB (glycogen branching enzyme), which are representative of two glycogen synthesis operons, and the gluconeogenic gene pckA (phosphoenolpyruvate carboxykinase), and it exhibited effects on cell size and surface (adherence) properties. The mutated gene was designated csrA for carbon storage regulator. Its effect on glycogen biosynthesis was mediated independently of cyclic AMP (cAMP), the cAMP receptor protein, and guanosine 3'-bisphosphate 5'-bisphosphate (ppGpp), which are positive regulators of glgC expression. A plasmid clone of the native csrA gene strongly inhibited glycogen accumulation and affected the ability of cells to utilize certain carbon sources for growth. Nucleotide sequence analysis, complementation experiments, and in vitro expression studies indicated that csrA encodes a 61-amino-acid polypeptide that inhibits glycogen biosynthesis. Computer-assisted data base searches failed to identify genes or proteins that are homologous with csrA or its gene product.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baecker P. A., Greenberg E., Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli 1,4-alpha-D-glucan:1,4-alpha-D-glucan 6-alpha-D-(1, 4-alpha-D-glucano)-transferase as deduced from the nucleotide sequence of the glg B gene. J Biol Chem. 1986 Jul 5;261(19):8738–8743. [PubMed] [Google Scholar]
- Bohannon D. E., Connell N., Keener J., Tormo A., Espinosa-Urgel M., Zambrano M. M., Kolter R. Stationary-phase-inducible "gearbox" promoters: differential effects of katF mutations and role of sigma 70. J Bacteriol. 1991 Jul;173(14):4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bridger W. A., Paranchych W. relA Gene control of bacterial glycogen synthesis. Can J Biochem. 1978 Jun;56(6):403–406. doi: 10.1139/o78-063. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Dietzler D. N., Leckie M. P., Magnani J. L., Sughrue M. J., Bergstein P. E., Sternheim W. L. Contribution of cyclic adenosine 3':5'-monophosphate to the regulation of bacterial glycogen synthesis in vivo. Effect of carbon source and cyclic adenosine 3':5'-monophosphate on the quantitative relationship between the rate of glycogen synthesis and the cellular concentrations of glucose 6-phosphate and fructose 1,6-diphosphate in Escherichia coli. J Biol Chem. 1979 Sep 10;254(17):8308–8317. [PubMed] [Google Scholar]
- Dietzler D. N., Leckie M. P. Regulation of ADP-glucose synthetase, the rate-limiting enzyme of bacterial glycogen synthesis, by the pleiotropic nucleotides ppGpp and pppGpp. Biochem Biophys Res Commun. 1977 Aug 22;77(4):1459–1467. doi: 10.1016/s0006-291x(77)80143-5. [DOI] [PubMed] [Google Scholar]
- Goldie H. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J Bacteriol. 1984 Sep;159(3):832–836. doi: 10.1128/jb.159.3.832-836.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot M. L., Chelm B. K. Isolation and expression of the Bradyrhizobium japonicum adenylate cyclase gene (cya) in Escherichia coli. J Bacteriol. 1984 Sep;159(3):1068–1071. doi: 10.1128/jb.159.3.1068-1071.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haziza C., Stragier P., Patte J. C. Nucleotide sequence of the asd gene of Escherichia coli: absence of a typical attenuation signal. EMBO J. 1982;1(3):379–384. doi: 10.1002/j.1460-2075.1982.tb01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R., Fischer D. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol. 1992 Jul;6(14):1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Leckie M. P., Ng R. H., Porter S. E., Compton D. R., Dietzler D. N. Regulation of bacterial glycogen synthesis. Stimulation of glycogen synthesis by endogenous and exogenous cyclic adenosine 3':5'-monophosphate in Escherichia coli and the requirement for a functional CRP gene. J Biol Chem. 1983 Mar 25;258(6):3813–3824. [PubMed] [Google Scholar]
- Leckie M. P., Tieber V. L., Porter S. E., Roth W. G., Dietzler D. N. Independence of cyclic AMP and relA gene stimulation of glycogen synthesis in intact Escherichia coli cells. J Bacteriol. 1985 Jan;161(1):133–140. doi: 10.1128/jb.161.1.133-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A., Auger E. A., Blum P. H., Schultz J. E. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- Okita T. W., Rodriguez R. L., Preiss J. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J Biol Chem. 1981 Jul 10;256(13):6944–6952. [PubMed] [Google Scholar]
- Petersen G. B., Stockwell P. A., Hill D. F. Messenger RNA recognition in Escherichia coli: a possible second site of interaction with 16S ribosomal RNA. EMBO J. 1988 Dec 1;7(12):3957–3962. doi: 10.1002/j.1460-2075.1988.tb03282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- Preiss J., Lammel C., Greenberg E. Biosynthesis of bacterial glycogen. Kinetic studies of a glucose-1-P adenylyltransferase (EC 2.7.7.27) from a glycogen-excess mutant of Escherichia coli B. Arch Biochem Biophys. 1976 May;174(1):105–119. doi: 10.1016/0003-9861(76)90329-5. [DOI] [PubMed] [Google Scholar]
- Preiss J., Romeo T. Physiology, biochemistry and genetics of bacterial glycogen synthesis. Adv Microb Physiol. 1989;30:183–238. doi: 10.1016/s0065-2911(08)60113-7. [DOI] [PubMed] [Google Scholar]
- Putney S. D., Royal N. J., Neuman de Vegvar H., Herlihy W. C., Biemann K., Schimmel P. Primary structure of a large aminoacyl-tRNA synthetase. Science. 1981 Sep 25;213(4515):1497–1501. doi: 10.1126/science.7025207. [DOI] [PubMed] [Google Scholar]
- Romeo T., Kumar A., Preiss J. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene. 1988 Oct 30;70(2):363–376. doi: 10.1016/0378-1119(88)90208-9. [DOI] [PubMed] [Google Scholar]
- Romeo T., Moore J., Smith J. A simple method for cloning genes involved in glucan biosynthesis: isolation of structural and regulatory genes for glycogen synthesis in Escherichia coli. Gene. 1991 Dec 1;108(1):23–29. doi: 10.1016/0378-1119(91)90483-r. [DOI] [PubMed] [Google Scholar]
- Romeo T., Preiss J. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5'-diphosphate 3'-diphosphate and analysis of in vivo transcripts. J Bacteriol. 1989 May;171(5):2773–2782. doi: 10.1128/jb.171.5.2773-2782.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Werner C., Ostell J., Tolstoshev C., Satterfield S. G. Mapping sequenced E.coli genes by computer: software, strategies and examples. Nucleic Acids Res. 1991 Feb 11;19(3):637–647. doi: 10.1093/nar/19.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellhorn H. E., Stones V. L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992 Jul;174(14):4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. Location of the maltose A and B loci on the genetic map of Escherichia coli. J Bacteriol. 1966 Oct;92(4):1083–1089. doi: 10.1128/jb.92.4.1083-1089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D. A., Kolter R. Life after log. J Bacteriol. 1992 Jan;174(2):345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Taguchi M., Izui K., Katsuki H. Augmentation of glycogen synthesis under stringent control in Escherichia coli. J Biochem. 1980 Aug;88(2):379–387. doi: 10.1093/oxfordjournals.jbchem.a132983. [DOI] [PubMed] [Google Scholar]
- Wanner B. L., Kodaira R., Neidhardt F. C. Physiological regulation of a decontrolled lac operon. J Bacteriol. 1977 Apr;130(1):212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991 Mar 25;266(9):5980–5990. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yu F., Jen Y., Takeuchi E., Inouye M., Nakayama H., Tagaya M., Fukui T. Alpha-glucan phosphorylase from Escherichia coli. Cloning of the gene, and purification and characterization of the protein. J Biol Chem. 1988 Sep 25;263(27):13706–13711. [PubMed] [Google Scholar]