Abstract
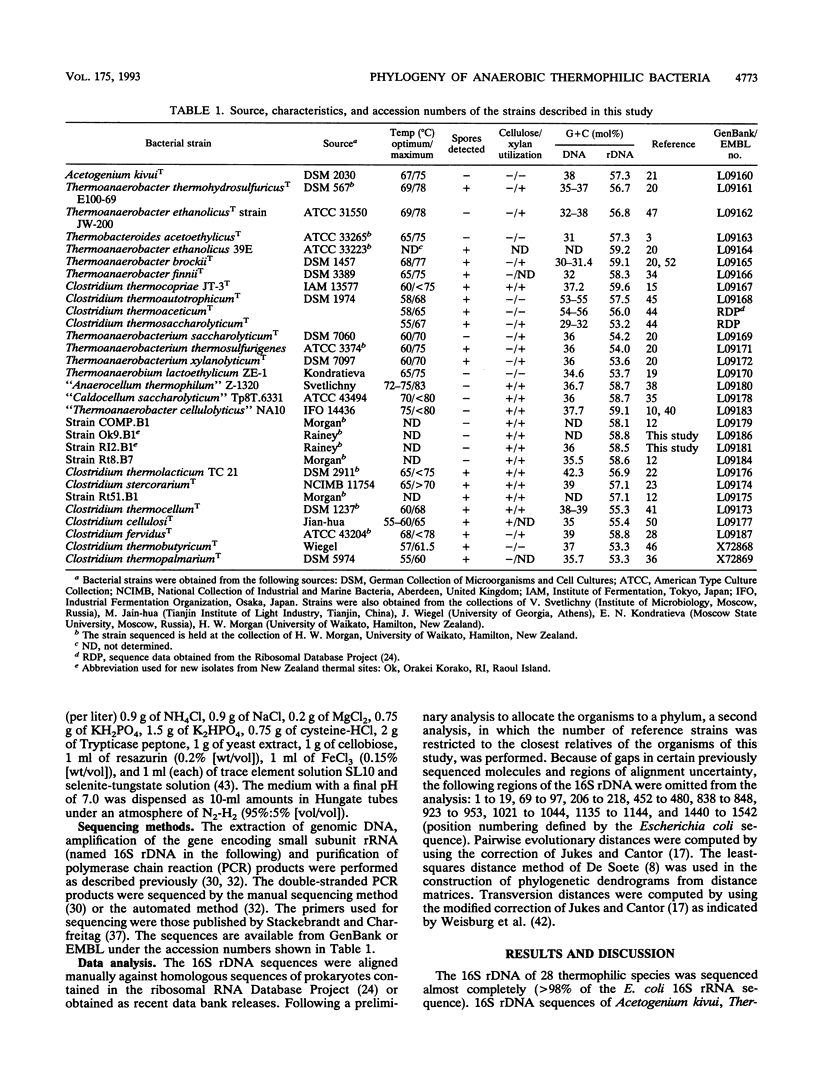
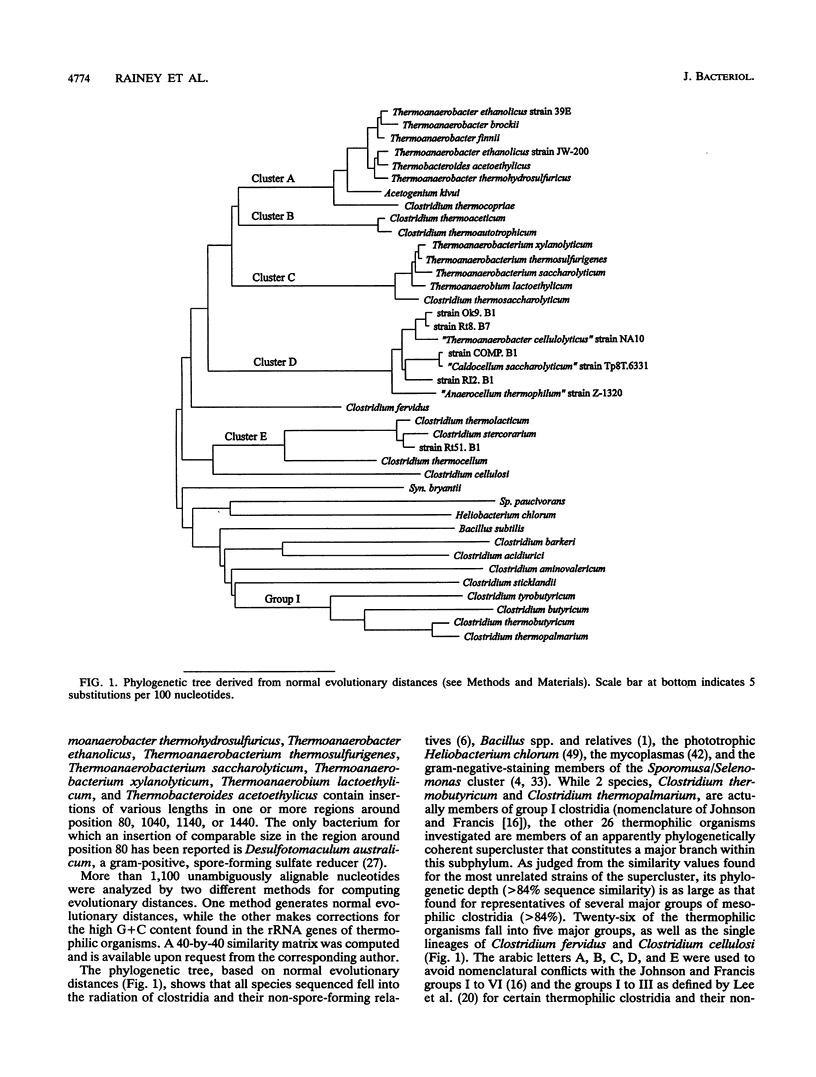
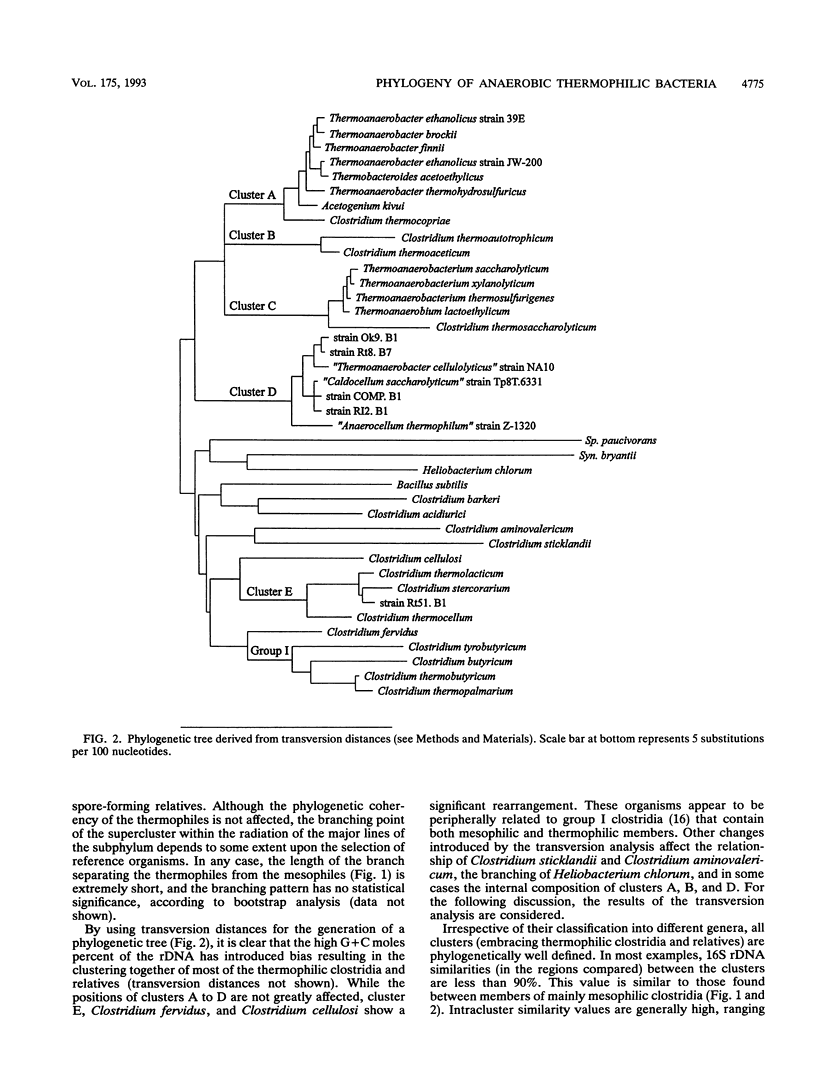
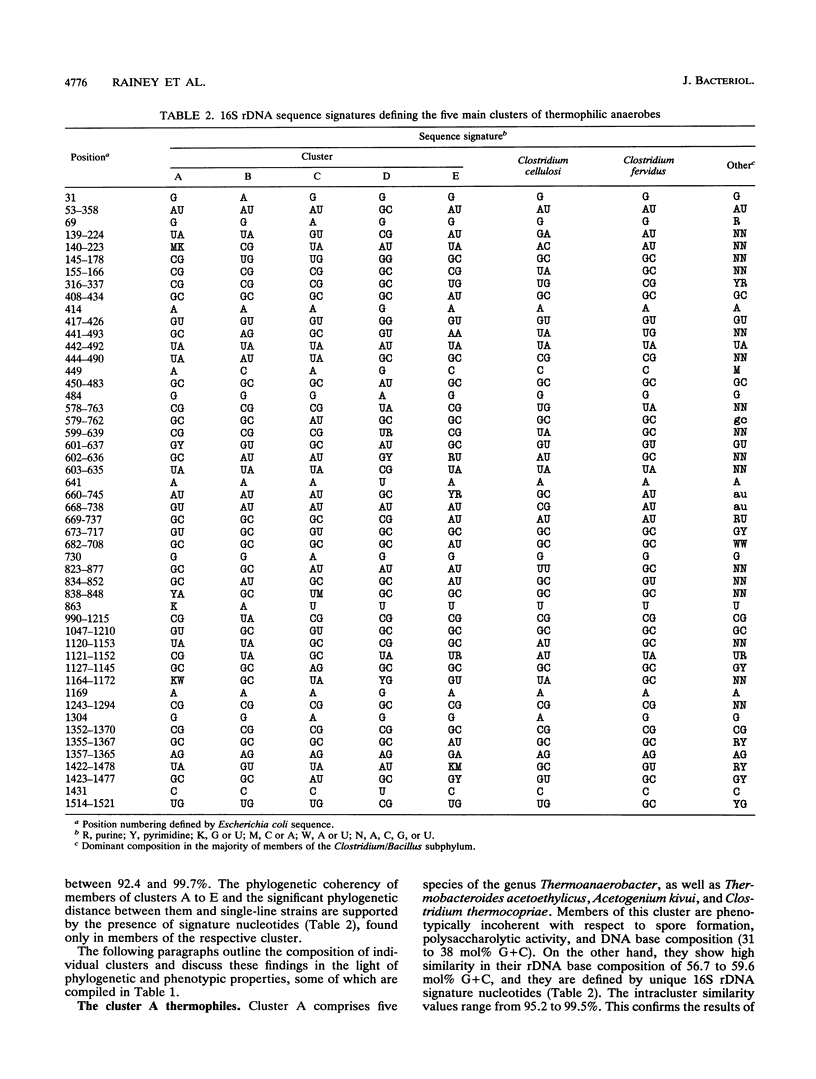
Small subunit rDNA sequences were determined for 20 species of the genera Acetogenium, Clostridium, Thermoanaerobacter, Thermoanaerobacterium, Thermoanaerobium, and Thermobacteroides, 3 non-validly described species, and 5 isolates of anaerobic thermophilic bacteria, providing a basis for a phylogenetic analysis of these organisms. Several species contain a version of the molecule significantly longer than that of Escherichia coli because of the presence of inserts. On the basis of normal evolutionary distances, the phylogenetic tree indicates that all bacteria investigated in this study with a maximum growth temperature above 65 degrees C form a supercluster within the subphylum of gram-positive bacteria that also contains Clostridium thermosaccharolyticum and Clostridium thermoaceticum, which have been previously sequenced. This supercluster appears to be equivalent in its phylogenetic depth to the supercluster of mesophilic clostridia and their nonspore-forming relatives. Several phylogenetically and phenotypically coherent clusters that are defined by sets of signature nucleotides emerge within the supercluster of thermophiles. Clostridium thermobutyricum and Clostridium thermopalmarium are members of Clostridium group I. A phylogenetic tree derived from transversion distances demonstrated the artificial clustering of some organisms with high rDNA G+C moles percent, i.e., Clostridium fervidus and the thermophilic, cellulolytic members of the genus Clostridium. The results of this study can be used as an aid for future taxonomic restructuring of anaerobic sporogenous and asporogenous thermophillic, gram-positive bacteria.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Both B., Buckel W., Kroppenstedt R., Stackebrandt E. Phylogenetic and chemotaxonomic characterization of Acidaminococcus fermentans. FEMS Microbiol Lett. 1992 Oct 1;76(1-2):7–11. doi: 10.1016/0378-1097(92)90355-r. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- He Y. L., Ding Y. F., Long Y. Q. Two cellulolytic Clostridium species: Clostridium cellulosi sp. nov. and Clostridium cellulofermentans sp. nov. Int J Syst Bacteriol. 1991 Apr;41(2):306–309. doi: 10.1099/00207713-41-2-306. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Francis B. S. Taxonomy of the Clostridia: ribosomal ribonucleic acid homologies among the species. J Gen Microbiol. 1975 Jun;88(2):229–244. doi: 10.1099/00221287-88-2-229. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyaizu H., Debrunner-Vossbrinck B., Mandelco L., Studier J. A., Woese C. R. The green non-sulfur bacteria: a deep branching in the eubacterial line of descent. Syst Appl Microbiol. 1987;9:47–53. doi: 10.1016/s0723-2020(87)80055-3. [DOI] [PubMed] [Google Scholar]
- Patel B. K., Love C. A., Stackebrandt E. Helix 6 of the 16S rRNA of the bacterium Desulfotomaculum australicum exhibits an unusual structural idiosyncrasy. Nucleic Acids Res. 1992 Oct 25;20(20):5483–5483. doi: 10.1093/nar/20.20.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissons C. H., Sharrock K. R., Daniel R. M., Morgan H. W. Isolation of cellulolytic anaerobic extreme thermophiles from new zealand thermal sites. Appl Environ Microbiol. 1987 Apr;53(4):832–838. doi: 10.1128/aem.53.4.832-838.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990 Jan;136(1):37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Debrunner-Vossbrinck B. A., Oyaizu H., Stackebrandt E., Ludwig W. Gram-positive bacteria: possible photosynthetic ancestry. Science. 1985;229:762–765. doi: 10.1126/science.11539659. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Ben-Bassat A., Hegge P. W. Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol. 1980 Jul;143(1):432–440. doi: 10.1128/jb.143.1.432-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]


