Abstract
We recently isolated an RNA polymerase II elongation factor from rat liver nuclei and found it to be homologous to the product of the human ELL gene, a frequent target for translocations in acute myeloid leukemia. To further our understanding of the possible role(s) of ELL in transcriptional regulation and human disease, we initiated a search for ELL-related proteins. In this report we describe molecular cloning, expression, and characterization of human ELL2, a novel RNA polymerase II elongation factor 49% identical and 66% similar to ELL. Mechanistic studies indicate that ELL2 and ELL possess similar transcriptional activities. Structure–function studies localize the ELL2 elongation activation domain to an ELL2 N-terminal region that is highly homologous to ELL. Finally, Northern blot analysis reveals that the ELL2 and ELL genes are transcribed in many of the same tissues, but that the ratio of their transcripts exhibits tissue-to-tissue variation, raising the possibility that ELL2 and ELL may not perform completely general functions, but, instead, may perform gene- or tissue-specific functions.
The elongation stage of eukaryotic messenger RNA synthesis is a major site for the regulation of gene expression (1, 2). Moreover, a growing body of evidence suggests that misregulation of transcription elongation may be a key element in a variety of human diseases (3).
To date, one virally encoded protein (Tat) and five cellular proteins [SII, P-TEFb, TFIIF, elongin (SIII), and ELL] have been defined biochemically and shown to be capable of controlling the activity of the RNA polymerase II elongation complex. Among these elongation factors, three have been implicated in human disease. The HIV-1 encoded Tat protein is required for efficient transcription of HIV-1 genes and for productive infection by the virus (4). Elongin (SIII) is a potential target for regulation by the product of the von Hippel–Lindau tumor suppressor gene, which is mutated in the majority of clear-cell renal carcinomas and in families with von Hippel–Lindau disease, a rare genetic disorder that predisposes individuals to a variety of cancers (5, 6). The ELL gene on chromosome 19p13.1 was originally isolated as a gene that undergoes frequent translocations with the Drosophila trithorax-like MLL gene on chromosome 11q23 in acute myeloid leukemia (7, 8).
As part of our effort to understand how elongation by RNA polymerase II is controlled under normal cell conditions and in disease, we are attempting to reconstitute the RNA polymerase II elongation apparatus. In this report, we describe identification and characterization of ELL2, a novel ELL-related RNA polymerase II elongation factor from human cells.
MATERIALS AND METHODS
Cloning and Expression of Wild-Type and Mutant ELL2.
Searches of the Human Genome Sciences and GenBank databases identified overlapping expressed sequence tags (ESTs) that form a contig spanning a predicted ELL2 ORF. These ESTs include Human Genome Sciences cDNAs HNEAK22, HNFDO55, HBWAL95R, HBWAH80R, HSBAI43R, HOUDO79R, HCE2D15R, HPRAE28R, PM1163767, HSXCR53RA, and HATDQ29R (9, 10) and GenBank cDNAs with accession numbers W94585W94585, T89063T89063, R16400R16400, R12663R12663, W92650W92650, AA009921, PM770010, and PM717576.
A DNA fragment including ELL2 coding sequences was obtained by PCR amplification of a Lambda Zap human fetal heart library using a 5′ primer (5′-CAATTAACCCTCATAAAGGGAAC-3′) identical to a sequence in the Lambda Zap vector and a 3′ antisense primer (5′-CAAAGTTTCACCTTTTAGAATCTAGAGCAACTC-3′) corresponding to a sequence in the 3′-untranslated region of the ELL2 gene. The construct for expression of histidine-tagged ELL2 in bacteria was prepared in two steps. First, a DNA fragment encoding ELL2 amino acids 11–640 was generated by PCR amplification of the original ELL2 ORF-containing PCR product using the ELL2- specific primers 5′-GAGGTGTCGACGAGGAGCAGCGCTATGGGCTGTCGTGCGGAC-3′ and 5′-GTGTGGATCCTCATCACTAGGACCATGACTCTGCTTGCTGTTG3′ and was introduced into the SalI and BamHI sites of M13 mpET (11). An expression vector containing the entire ELL2 ORF was then generated by oligonucleotide-directed mutagenesis (12) with the Muta-Gene M13 in vitro mutagenesis kit (Bio-Rad) and confirmed by DNA sequencing; N- and C-terminal ELL2 deletion mutants were constructed by the same procedure. Wild-type and mutant ELL2 proteins were expressed in Escherichia coli, purified from guanidine-solubilized inclusion bodies by nickel affinity chromatography, and renatured as described (13). Where indicated, the ELL2 protein was further purified by preparative SDS/PAGE (13). The human ELL protein was expressed in E. coli and purified as described (13).
RESULTS
Identification of Human ELL2.
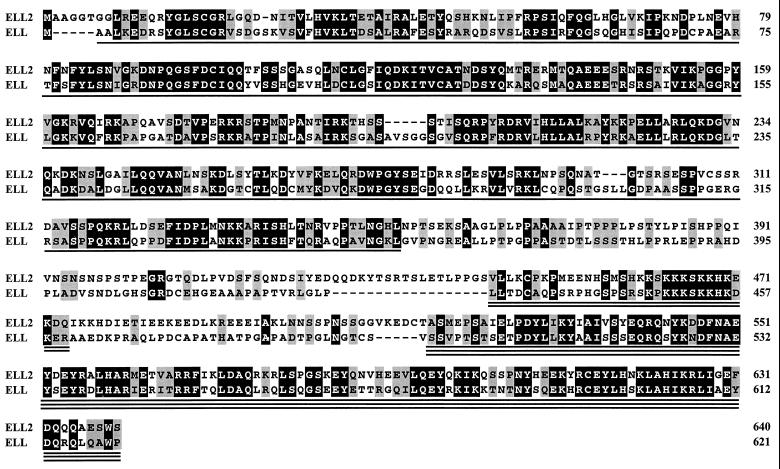
Searches of the Human Genome Sciences and GenBank databases identified multiple overlapping ESTs that formed a contig spanning a predicted ELL2 ORF similar in sequence to the ORF of the human ELL gene (Fig. 1). An ≈1.9-kb DNA fragment containing the entire predicted ELL2 ORF was obtained by PCR amplification of a human fetal heart library and sequenced. The ELL2 ORF encodes a 640-amino acid protein with a calculated molecular mass of 72,354 Da. As determined by the bestfit program of the Genetics Computer Group (Madison, WI) package (14), ELL2 is 49% identical and 66% similar to ELL (alignment score ≈64 SD).
Figure 1.
Comparison of the deduced amino acid sequences of human ELL2 and ELL. ELL2 residues 11–640 are from the sequence of a DNA fragment obtained by PCR amplification of a human fetal heart cDNA library. Residues 1–10 are from Human Genome Sciences ESTs HNFD055 and HNEAK22 and from an EST in the GenBank database (accession number W94585W94585). Identical amino acids are shown in white letters on a black background; similar amino acids (A, S, T, P; D, E, N, Q; H, R, K; I, L, M, V; F, Y, W) are shown in black letters on a gray background. Conserved regions 1, 2, and 3 are indicated by single, double, and triple underlines, respectively.
Expression of ELL2 and ELL in Human Cells.
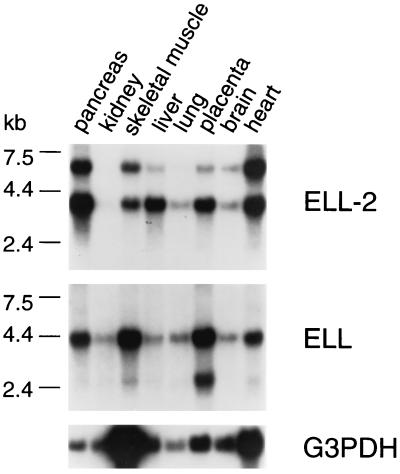
To investigate the expression of ELL2 and ELL in human tissues, Northern blots containing poly(A)+ RNA from various human tissues were hybridized with ELL2- and ELL-specific probes (Fig. 2). Consistent with previous studies (7, 8), the ELL-specific probe hybridized to two mRNA species of ≈4.4 kb and ≈2.7 kb. As shown in Fig. 2, the ELL2-specific probe hybridized to two mRNA species of ≈6 kb and ≈4.1 kb. At present, it is not clear whether the ≈6-kb and ≈4.1-kb ELL2 mRNAs are alternatively processed forms or the products of closely related genes. The results of Northern blot analyses indicate that both ELL2 and ELL mRNAs are expressed in many of the same tissues. Notably, the ratio of ELL2 and ELL mRNAs, and the ratios of the two different forms of each mRNA, exhibit tissue-to-tissue variation; for example, the ratio of ELL2 to ELL is greater in liver than in kidney, skeletal muscle, lung, and placenta.
Figure 2.
Tissue distribution of ELL2 and ELL mRNAs. A human multiple tissue northern blot (MTN1, CLONTECH) was probed sequentially with PCR-generated ELL2- and ELL-specific probes chosen from a region of sequence that was most divergent between the two genes. The ELL-specific probe contained sequences encoding amino acids 317–621, and the ELL2-specific probe contained sequences encoding amino acids 327–474. As a loading control, the same blot was probed with a probe specific for glyceraldehyde-3-phosphate dehydrogenase (G3PDH) mRNA. Probes were labeled with [α-32P]dCTP by random priming performed according to the manufacturer’s instructions (Rediprime kit, Amersham). The blot was prehybridized in 10 ml of Hybrisol I solution (Oncor) for 3 h at 42°C. Probe DNA was denatured and added to hybridization solution at 106 cpm/ml of solution. Hybridization was carried out at 42°C overnight. The blot was washed 10 min in 2× standard saline citrate (SSC)/0.1% SDS at room temperature, 15 min in 0.2× SSC/0.1% SDS at 45°C, 10 min in 0.1× SSC/0.1% SDS at 55°C, and then exposed to film (Hyperfilm-MP, Amersham) overnight at −80°C.
ELL2 and ELL Possess Similar Transcriptional Activities.
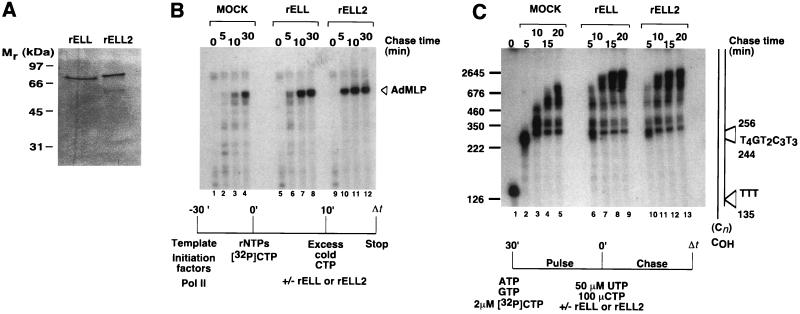
In a previous study, we demonstrated that ELL is capable of potently stimulating the overall rate of RNA chain elongation by RNA polymerase II (13). To determine whether ELL2 is also capable of stimulating elongation by RNA polymerase II, a DNA fragment containing the ELL2 ORF was introduced into a bacteriophage M13 expression vector under control of the T7 RNA polymerase promoter (11) and expressed in E. coli with an N-terminal histidine tag. The recombinant ELL2 protein was purified to homogeneity from guanidine-solubilized inclusion bodies by nickel-affinity chromatography and preparative SDS/PAGE (Fig. 3A) and then tested for its ability to stimulate elongation.
Figure 3.
ELL2 and ELL have similar effects on elongation by RNA polymerase II during synthesis of promoter-independent and promoter-dependent transcripts. (A) Ten percent SDS/PAGE of recombinant ELL2 (rELL2) and ELL (rELL), purified by nickel chromatography and preparative SDS/PAGE. Proteins were visualized by silver staining. (B) Effects of ELL2 and ELL on the kinetics of promoter-dependent transcription. Preinitiation complexes were assembled at the AdML promoter with recombinant TBP, TFIIB, TFIIE, TFIIF, and purified rat TFIIH and RNA polymerase II as described (13). Transcription was initiated by addition of 50 μM ATP, 50 μM GTP, 2 μM UTP, 10 μCi of [α-32P]CTP (>400 Ci/mmol, Amersham), and 7 mM MgCl2. After 10 min at 28°C, 100 μM nonradioactive CTP was added to reaction mixture and short transcripts were chased in the absence or presence of ≈50 ng of SDS/PAGE-purified rELL2 or rELL for the times indicated. Transcripts were analyzed by electrophoresis through a 6% polyacrylamide/7.0 M urea gel. (C) Effects of ELL2 and ELL on the kinetics of promoter-independent transcription. SDS/PAGE-purified histidine-tagged ELL2 and ELL proteins were renatured and assayed in pulse–chase reactions as diagrammed in the figure using the oligo(dC)-tailed template pCpGR220 S/P/X. Reactions contained ≈0.01 unit of RNA polymerase II, 100 ng of pCpGR220S/P/X, and ≈50 ng of rELL2 or ≈50 ng of ELL and were performed essentially as described (13). The control reaction (mock) contained an identically prepared fraction from uninfected JM109(DE3) cells.
As shown in Figs. 3 B and C, ELL2 is an RNA polymerase II elongation factor with functional properties similar to those of ELL. In these experiments, the abilities of ELL2 and ELL to stimulate elongation were compared during either promoter-specific transcription carried out in the presence of the general initiation factors or promoter-independent transcription carried out using an oligo(dC)-tailed template assay, in the absence of auxillary transcription factors.
To compare the abilities of ELL2 and ELL to stimulate the rate of elongation of promoter-specific transcripts, preinitiation complexes were assembled by preincubation of purified RNA polymerase II, TBP, TFIIB, TFIIE, TFIIF, and TFIIH with a DNA template containing the AdML promoter. Short, highly radioactive transcripts were then synthesized during a brief pulse carried out in the presence of ATP, GTP, UTP, and a limiting concentration of [α-32P]CTP. These short, promoter-specific transcripts were then chased into full-length runoff transcripts in the presence of an excess of nonradioactive CTP and in the presence or absence of approximately equivalent levels of recombinant ELL2 or ELL. As shown in Fig. 3B, comparison of the kinetics of accumulation of full-length runoff transcripts reveals that ELL2 and ELL have similar effects on the rate of elongation of promoter-specific transcripts by RNA polymerase II.
An oligo(dC)-tailed template assay was used to compare the abilities of ELL2 and ELL to stimulate the rate of elongation of promoter-independent transcripts. Briefly, transcription was initiated by addition of RNA polymerase II to reaction mixtures containing the oligo(dC)-tailed template pCpGR220 S/P/X (15), ATP, GTP, and [α-32P]CTP. Under these conditions, RNA polymerase II synthesizes ≈135-nucleotide transcripts on the T-less cassette of pCpGR220 S/P/X. These highly radioactive transcripts were then chased into longer RNAs with UTP and an excess of nonradioactive CTP, in the presence or absence of approximately equivalent levels of recombinant ELL2 or ELL. As shown in Fig. 3C, transcripts synthesized in the presence of either ELL2 or ELL were substantially longer than transcripts synthesized in their absence; we note that many transcripts synthesized in the presence of ELL2 and ELL appear to be plasmid length. In addition, comparison of the kinetics of accumulation of long transcripts and of the distribution of RNA intermediates reveals that ELL2 and ELL have similar effects on elongation of transcripts synthesized by RNA polymerase II in the absence of auxillary transcription factors on the oligo(dC)-tailed pCpGR220 S/P/X template.
Localization of the ELL2 Elongation Activation Domain.
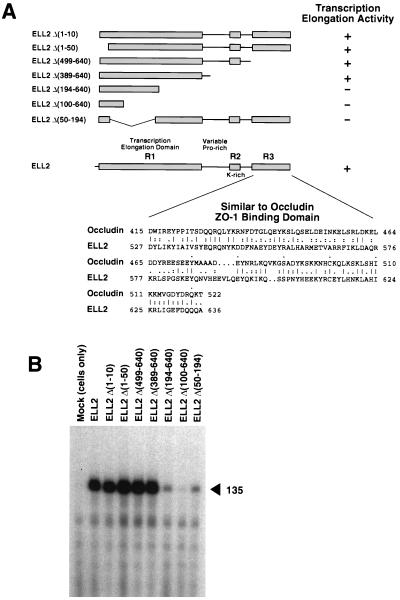
Comparison of the ELL2 and ELL ORFs revealed three conserved regions (Figs. 1 and 4A): an N-terminal region (region 1, R1) between ELL2 residues 7 and 350, a short lysine-rich region (region 2, R2) between ELL2 residues 443 and 474, and a C-terminal region (region 3, R3) between ELL2 residues 516 and 640. Although neither ELL2 nor ELL have obvious structural features such as zinc finger, leucine zipper, or helix-turn-helix motifs commonly found in transcription factors, a tblastn search of the GenBank database revealed that conserved region 3 of ELL2 and ELL exhibits striking similarity to the ZO-1 binding domain of occludin (16), an integral membrane protein found at tight junctions (17). As determined by the bestfit program of the Genetics Computer Group package (14), the C terminus of ELL2 and the ZO-1 binding domain of occludin are 33% identical and 61% similar (alignment score ≈17 SD) over a 112-amino acid region. In addition, ELL2 and ELL each contain a proline-rich, nonconserved region that bridges conserved regions 1 and 2. The ELL2 proline-rich region includes several PXXP motifs that are potential binding sites for SH3 domains (18).
Figure 4.
Localization of the ELL2 elongation activation domain. (A) Summary of ELL2 mutants and their activities in transcription. Wild-type ELL2 is diagrammed at the bottom of the panel. Conserved regions 1, 2, and 3 (R1, R2, and R3) are indicated by the shaded boxes. The alignment of region 3 with the C-terminal ZO-1 binding domain of occludin was generated with the bestfit program of the Genetics Computer Group package, using the symbol comparison table of Gribskov and Burgess (34). (B) Wild-type ELL2 and ELL2 mutants were expressed in E. coli and purified by nickel-affinity chromatography as described (13). Approximately 50 ng of each protein (in a maximum volume of 50 μl) was renatured and assayed as described (13) for its ability to stimulate synthesis of the 135-nucleotide transcript from the T-less cassette of oligo(dC)-tailed template pCpGR220 S/P/X. Reactions containing ≈0.01 unit of RNA polymerase II, 100 ng template, and the indicated ELL2 proteins were incubated at 28°C for 5 min in the presence of 50 μM ATP, 50 μM GTP, 1.8 μM CTP, and 10 μCi of [α-32P]CTP. The control reaction (MOCK) contained an identically prepared fraction from uninfected JM109(DE3) cells.
To assess the functional significance of the regions conserved between ELL2 and ELL and to localize the ELL2 elongation activation domain, a series of ELL2 deletion mutants was constructed (Fig. 4A), expressed in E. coli, purified, and tested for transcriptional activity using the oligo(dC)-tailed template assay. As shown in Fig. 4B, ELL2 deletion mutants Δ194–640, Δ100–640, and Δ50–194, which each lack significant portions of region 1, had significantly reduced transcriptional activities. In contrast, ELL2 deletion mutants Δ1–10, Δ1–50, Δ499–640, and Δ389–640 all exhibited near wild-type levels of activity. These results demonstrate that regions 2 and 3 are dispensable for ELL2 elongation activity and that sequences in conserved region 1 between residues 50 and 389 are sufficient for maximal ELL2 elongation activity and therefore include the ELL2 elongation activation domain. At the present time, we do not know whether the elongation activation domain includes the entire region from residues 50 to 389 or whether some portion of this region is required for proper folding of the protein during solubilization from inclusion bodies.
DISCUSSION
Here we report identification and characterization of ELL2, a novel RNA polymerase II elongation factor similar to previously characterized elongation factor ELL (13). ELL2 is the newest addition to a growing list of biochemically defined cellular proteins that are capable of regulating the activity of the RNA polymerase II elongation complex. In addition to proteins such as HMG-14 and SWI/SNF-like complexes, which appear to affect elongation by altering chromatin structure (19, 20), this list includes the six cellular elongation factors SII, P-TEFb, TFIIF, Elongin (SIII), ELL, and ELL2, which act directly on the ternary elongation complex and fall into two distinct functional classes (1).
SII and P-TEFb were shown previously to prevent RNA polymerase II from arresting transcription prematurely. SII protects RNA polymerase II from arrest at a variety of transcriptional impediments, including specific DNA sequences that act as intrinsic arrest sites and some DNA-bound proteins and drugs. SII promotes passage of RNA polymerase II through these transcriptional impediments by a mechanism involving reiterative endonucleolytic cleavage and re-extension of nascent transcripts held in the polymerase site (21). P-TEFb promotes passage of RNA polymerase II through DRB-sensitive arrest sites within a few hundred nucleotides of promoters, by a mechanism that may involve phosphorylation of the RNA polymerase II CTD (22, 23). TFIIF, Elongin (SIII), and ELL were all shown previously to increase the overall rate of elongation by RNA polymerase II by decreasing the frequency or duration of transient pausing by the enzyme at many sites along DNA templates (13, 24, 25). Neither TFIIF, Elongin (SIII), nor ELL is capable of releasing RNA polymerase II from SII- or DRB-sensitive arrest sites.
As we have shown here, ELL2 regulates the activity of the RNA polymerase II elongation complex by a mechanism more closely resembling those of TFIIF, Elongin (SIII), and ELL than those of SII and P-TEFb. ELL2 appears to increase the overall rate of elongation by RNA polymerase II during both promoter-dependent and -independent transcription. In addition, we observe that, unlike SII, ELL2 does not release RNA polymerase II from arrest or promote the nascent transcript cleavage reaction (A.S., J.W.C., R.C.C., and J. Elmendorf, unpublished results).
Although ELL2 and ELL are related proteins, they do not share sequence similarity throughout their entire ORFs. Alignment of their ORFs revealed that ELL2 and ELL share three regions of high homology: an N-terminal region between ELL2 residues 7 and 353, a short lysine-rich region between ELL2 residues 443–474, and a C-terminal region between ELL2 residues 516–640. Structure-function analysis reveals that ELL2 transcriptional activity resides in conserved region 1 in the ELL2 N terminus. Neither the conserved lysine-rich region 2 nor the conserved C-terminal region 3 is required for ELL2 transcriptional activity. The functions of regions 2 and 3 are presently unknown.
A homology search of the GenBank database revealed that conserved region 3 of ELL2 and ELL bears a striking resemblance to the ZO-1 binding domain of occludin (16), an integral membrane protein localized at tight junctions in mammalian cells (17). ZO-1 is a member of the family of membrane-associated guanylate kinase homologs (MAGUKs) believed to be important in signal transduction originating from sites of cell-cell contact (26). The founding member of the MAGUK family of putative signaling proteins is the product of the lethal (1) discs large-1 (dlg) tumor suppressor gene of Drosophila (27). Other members of the MAGUK family include ZO-2, a second tight junction protein (28), PSD-95/SAP-90, which localizes to synaptic junctions (29), p55, which participates in erythrocyte membrane-cytoskeletal interactions (30), and hdlg, a human homolog of Drosophila dlg (31). Recently, ZO-1, which is found exclusively in the cytosol of contact-inhibited cultured cells, was found to translocate to the nucleus in subconfluent cells, suggesting that ZO-1 is involved in signaling pathways controlled by cell-cell contact (32). Intriguingly, the intracellular localization of the product of the von Hippel–Lindau tumor suppressor gene, which has been shown to interact with and negatively regulate the B and C regulatory subunits of elongin, is similarly regulated by cell density (33). Whether conserved region 3 of ELL2 or ELL is capable of interacting with ZO-1 is presently unknown. It is tempting to speculate, however, that ELL2 and ELL could be regulated via a signal transduction pathway involving ZO-1 or ZO-1-like protein(s).
Finally, because of their abilities to stimulate elongation by RNA polymerase II through a wide variety of DNA template sequences, TFIIF, Elongin (SIII), and ELL have been considered “general” transcription elongation factors. Our finding that the ELL2 and ELL genes are expressed in many of the same tissues, but that the ratio of ELL2 and ELL mRNAs exhibits tissue-to-tissue variation, raises the possibility that ELL2 and ELL may perform gene- or tissue-specific functions. Future studies investigating this possibility will be crucial for an understanding of the roles ELL2, ELL, and the remaining elongation factors play in gene regulation.
Acknowledgments
We thank William Tuttle for technical assistance, Kenneth Jackson of the Molecular Biology Resource Center at the Oklahoma Center for Molecular Medicine for oligonucleotide synthesis, the Human Genome Sciences sequencing group for cDNA sequencing, and the National Disease Registry Institute for human tissues. This work was supported by National Institutes of Health Grant GM41628 (R.C.C. and J.W.C.), by a Merit Review Grant from the Department of Veterans’ Affairs (W.H.S.), and by funds provided to the Oklahoma Medical Research Foundation by the H. A. and Mary K. Chapman Charitable Trust. A.S. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
ABBREVIATION
- EST
expressed sequence tag
Footnotes
References
- 1.Reines D, Conaway J W, Conaway R C. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley D L. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 3.Aso T, Shilatifard A, Conaway J W, Conaway R C. J Clin Invest. 1996;97:1561–1569. doi: 10.1172/JCI118580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 5.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 6.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 7.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitani K, Kanda Y, Ogawa S, Tanaka T, Inazawa J, Yazaki Y, Hirai H. Blood. 1995;85:2017–2024. [PubMed] [Google Scholar]
- 9.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, Kerlavage A R, McCombie W R, Venter J C. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 10.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 11.Tan S, Conaway R C, Conaway J W. BioTechniques. 1994;16:824–828. [PubMed] [Google Scholar]
- 12.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 14.Genetics Computer Group. Program Manual for the GCG Package, Version 8. WI: Madison; 1994. [Google Scholar]
- 15.Rice G A, Kane C M, Chamberlin M J. Proc Natl Acad Sci USA. 1991;88:4245–4249. doi: 10.1073/pnas.88.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukito S. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, Chen J K, Feng S, Dalgarno D, Brauer A W, Schreiber S L. Cell. 1996;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 19.Ding H F, Rimsky S, Batson S C, Bustin M, Hansen U. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 20.Brown S A, Imbalzano A N, Kingston R E. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 21.Reines D. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. New York: Raven; 1994. pp. 263–278. [Google Scholar]
- 22.Marshall N F, Price D H. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 23.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 24.Price D H, Sluder A E, Greenleaf A L. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aso T, Lane W S, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 26.Willott E, Balda M S, Fanning A S, Jameson B, van Itallie C, Anderson J M. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods D F, Bryant P J. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 28.Jesaitis L A, Goodenough D A. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kistner U, Wenzel B M, Veh R W, Cases-Langhoff C, Garner A M, Appeltauer U, Voss B, Gundelfinger E D, Garner C C. J Biol Chem. 1993;268:4580–4583. [PubMed] [Google Scholar]
- 30.Ruff P, Speicher D W, Husain-Chishti A. Proc Natl Acad Sci USA. 1991;88:6595–6599. doi: 10.1073/pnas.88.15.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lue R A, Marfatia S M, Branton D, Chishti A H. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottardi C J, Arpin M, Fanning A S, Louvard D. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gribskov M, Burgess R R. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]