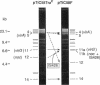
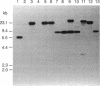
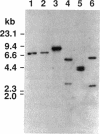
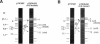Abstract
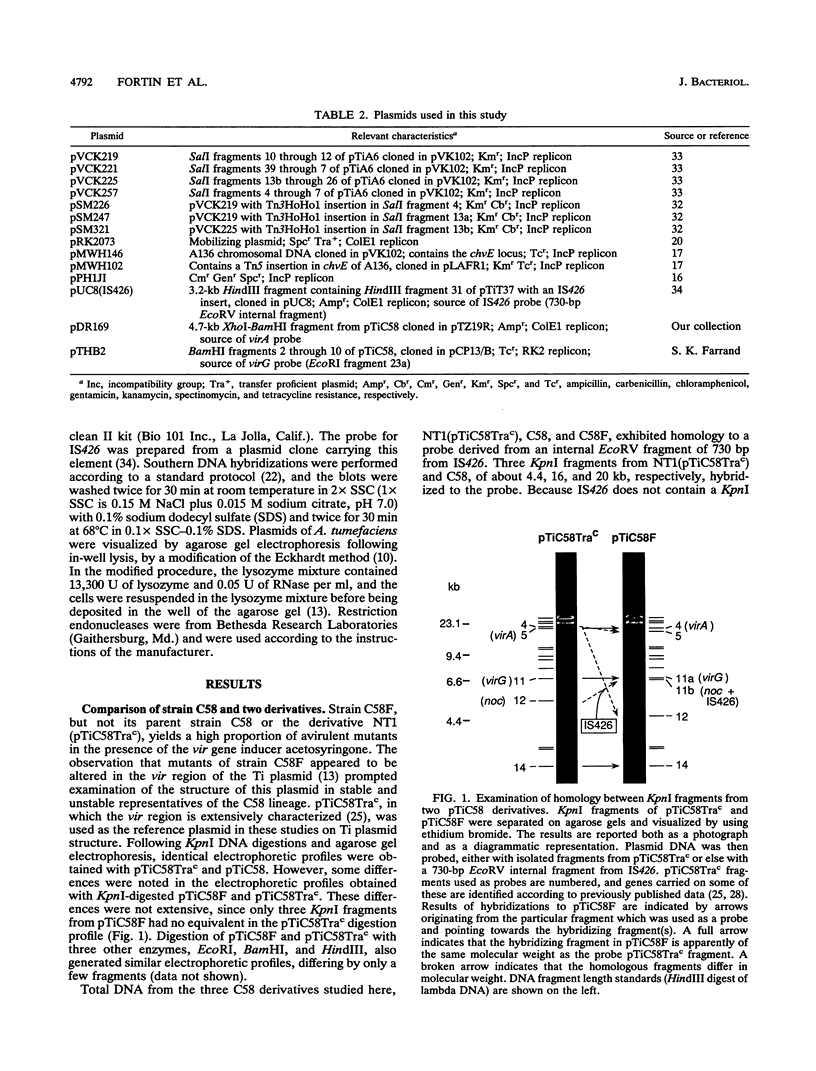
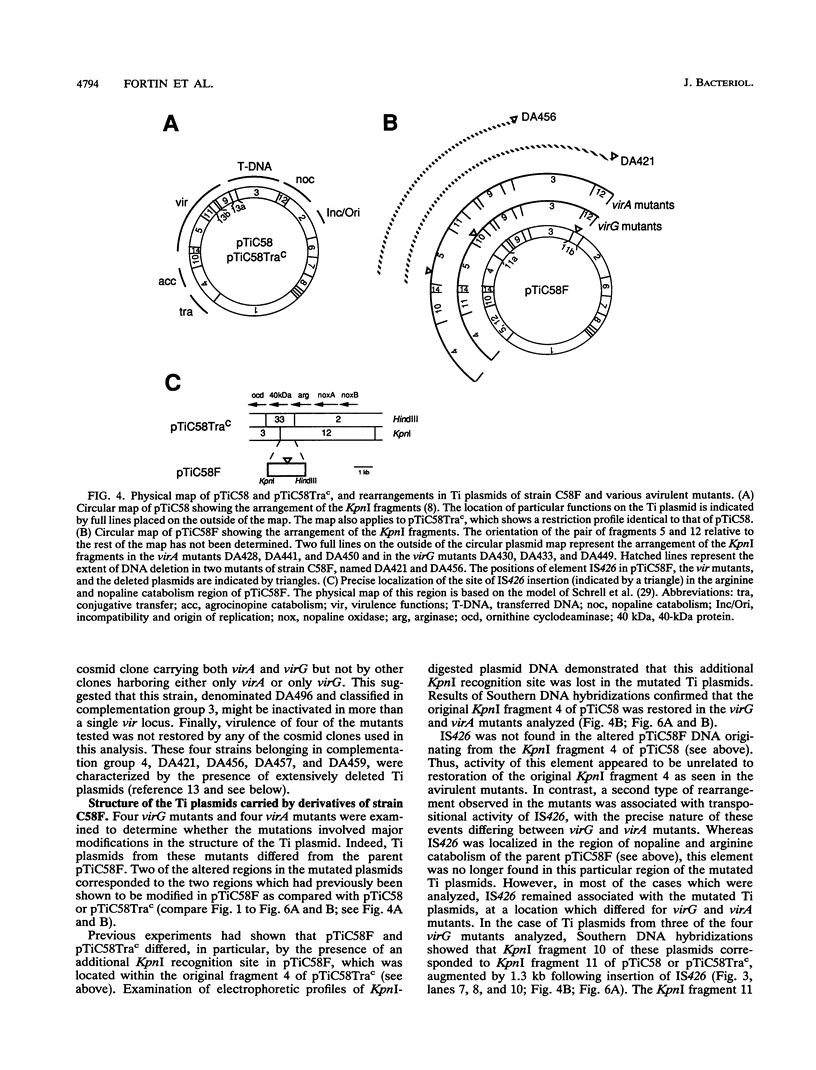
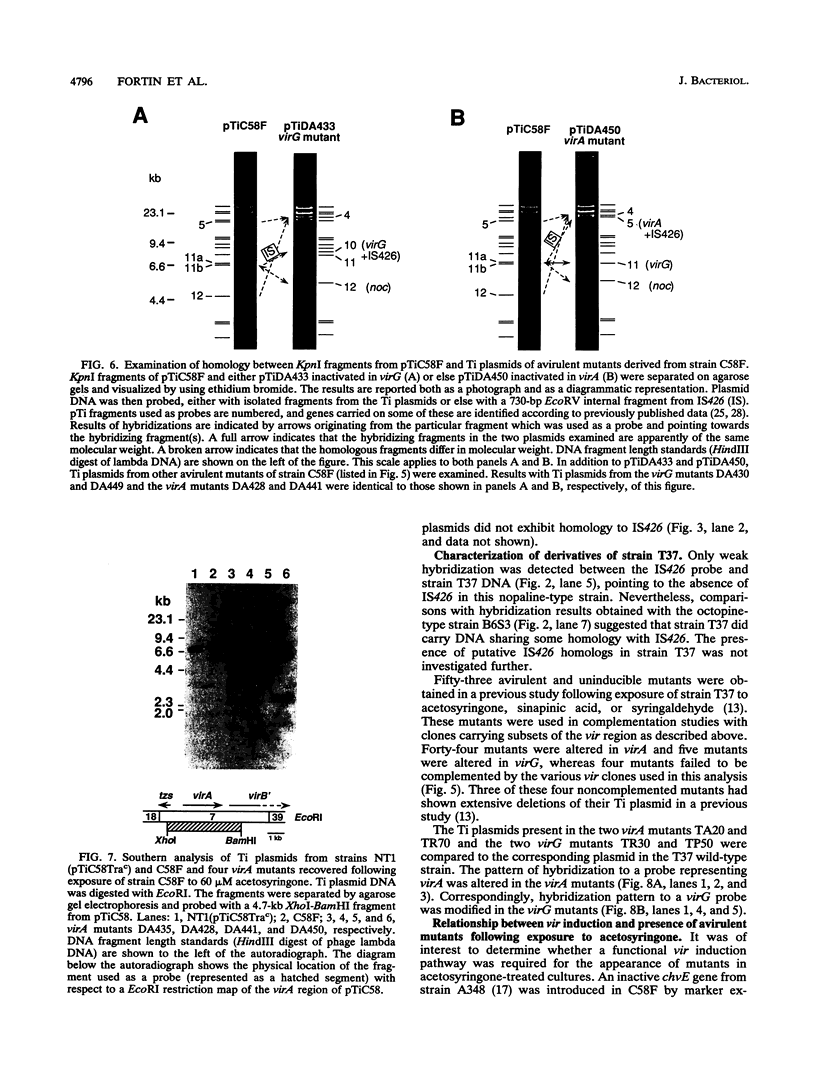
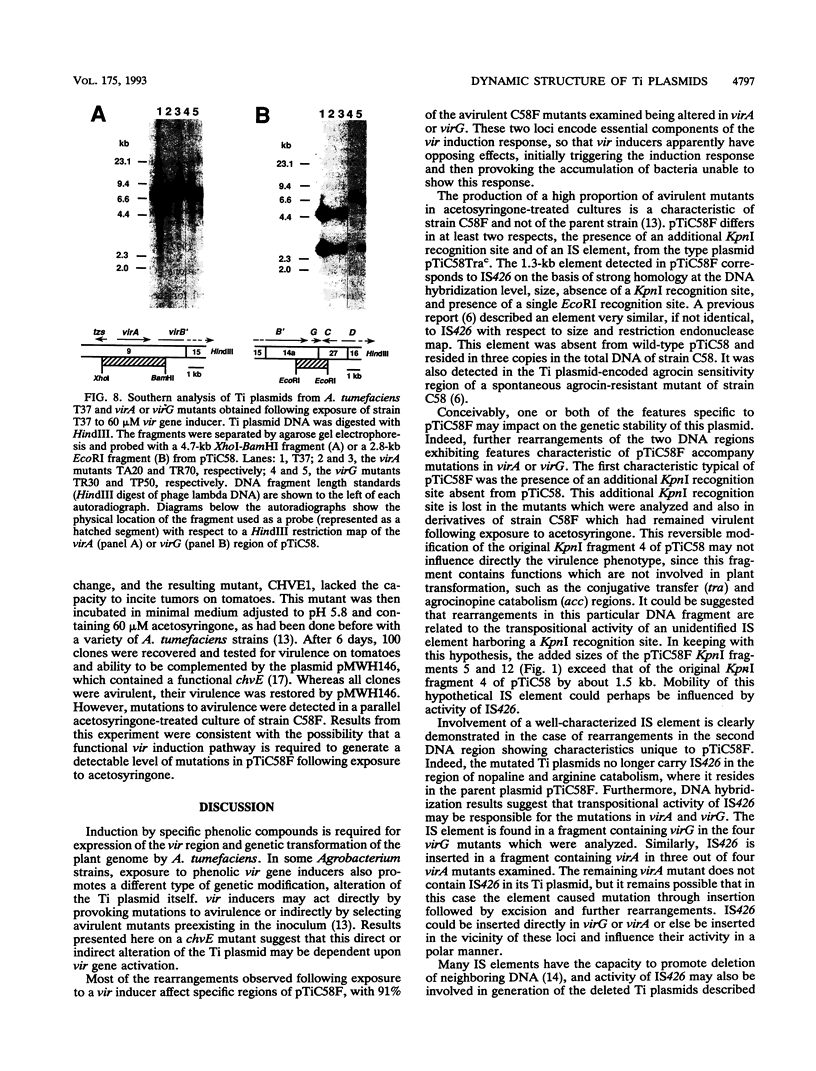
Agrobacterium tumefaciens C58F is a variant of strain C58 which generates a high proportion of avirulent mutants in the presence of the virulence (vir) gene inducer acetosyringone. These mutants are altered in the Ti plasmid and do not respond to the acetosyringone signal (C. Fortin, E. W. Nester, and P. Dion, J. Bacteriol. 174:5676-5685, 1992). The physical organization of the Ti plasmid was compared in strain C58 and its variant. One feature distinguishing pTiC58F from its parent plasmid was the presence of the insertion element IS426. Three copies of this element were detected in the strain C58 chromosome, whereas two additional copies were found in strain C58F, including one copy in the Ti plasmid. This particular copy of IS426 was associated with the region of arginine and nopaline catabolism of pTiC58F. Most of the avirulent mutants recovered following growth of strain C58F in the presence of acetosyringone were complemented by clones carrying either virA or virG. Element IS426 was no longer found in the arginine and nopaline catabolism region of the Ti plasmids from the virA and virG mutants, but it resided in the particular KpnI fragment containing the modified vir locus. Behavior of a strain C58F derivative, which was inactivated in a chromosomal component required for the response to acetosyringone, was consistent with the possibility that vir gene induction is essential to the massive production of avirulent mutants.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binns A. N., Sciaky D., Wood H. N. Variation in hormone autonomy and regenerative potential of cells transformed by strain A66 of Agrobacterium tumefaciens. Cell. 1982 Dec;31(3 Pt 2):605–612. doi: 10.1016/0092-8674(82)90316-6. [DOI] [PubMed] [Google Scholar]
- Chilton M. D., Currier T. C., Farrand S. K., Bendich A. J., Gordon M. P., Nester E. W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare B. G., Kerr A., Jones D. A. Characteristics of the nopaline catabolic plasmid in Agrobacterium strains K84 and K1026 used for biological control of crown gall disease. Plasmid. 1990 Mar;23(2):126–137. doi: 10.1016/0147-619x(90)90031-7. [DOI] [PubMed] [Google Scholar]
- Cooksey D. A. A spontaneous insertion in the agrocin sensitivity region of the Ti-plasmid of Agrobacterium tumefaciens C58. Plasmid. 1986 Nov;16(3):222–224. doi: 10.1016/0147-619x(86)90061-2. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Depicker A., De Wilde M., De Vos G., De Vos R., Van Montagu M., Schell J. Molecular cloning of overlapping segments of the nopaline Ti-plasmid pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980 Mar;3(2):193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Farrand S. K., Tempé J., Dessaux Y. Localization and characterization of the region encoding catabolism of mannopinic acid from the octopine-type Ti plasmid pTi15955. Mol Plant Microbe Interact. 1990 Jul-Aug;3(4):259–267. doi: 10.1094/mpmi-3-259. [DOI] [PubMed] [Google Scholar]
- Farrand S. K., Wang C. L., Hong S. B., O'Morchoe S. B., Slota J. E. Deletion derivatives of pAgK84 and their use in the analysis of Agrobacterium plasmid functions. Plasmid. 1992 Nov;28(3):201–212. doi: 10.1016/0147-619x(92)90052-c. [DOI] [PubMed] [Google Scholar]
- Fortin C., Nester E. W., Dion P. Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J Bacteriol. 1992 Sep;174(17):5676–5685. doi: 10.1128/jb.174.17.5676-5685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Simpson R. B., Ream L. W., White F. F., Gordon M. P., Nester E. W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981 Nov;27(1 Pt 2):143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- Hirsch P. R., Beringer J. E. A physical map of pPH1JI and pJB4JI. Plasmid. 1984 Sep;12(2):139–141. doi: 10.1016/0147-619x(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Huang M. L., Cangelosi G. A., Halperin W., Nester E. W. A chromosomal Agrobacterium tumefaciens gene required for effective plant signal transduction. J Bacteriol. 1990 Apr;172(4):1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J. C., Perry K. L., Kado C. I. Indoleacetic acid complementation and its relation to host range specifying genes on the Ti plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188(3):425–432. doi: 10.1007/BF00330044. [DOI] [PubMed] [Google Scholar]
- Leong S. A., Ditta G. S., Helinski D. R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982 Aug 10;257(15):8724–8730. [PubMed] [Google Scholar]
- Machida Y., Sakurai M., Kiyokawa S., Ubasawa A., Suzuki Y., Ikeda J. E. Nucleotide sequence of the insertion sequence found in the T-DNA region of mutant Ti plasmid pTiA66 and distribution of its homologues in octopine Ti plasmid. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7495–7499. doi: 10.1073/pnas.81.23.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten L., Canaday J., Gérard J. C., Fournier P., Crouzet P., Paulus F. Evolution of agrobacteria and their Ti plasmids--a review. Mol Plant Microbe Interact. 1992 Jul-Aug;5(4):279–287. doi: 10.1094/mpmi-5-279. [DOI] [PubMed] [Google Scholar]
- Rogowsky P. M., Powell B. S., Shirasu K., Lin T. S., Morel P., Zyprian E. M., Steck T. R., Kado C. I. Molecular characterization of the vir regulon of Agrobacterium tumefaciens: complete nucleotide sequence and gene organization of the 28.63-kbp regulon cloned as a single unit. Plasmid. 1990 Mar;23(2):85–106. doi: 10.1016/0147-619x(90)90028-b. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Long S. R., Meade H. M., van den Bos R. C., Ausubel F. M. ISRm1: A Rhizobium meliloti insertion sequence that transposes preferentially into nitrogen fixation genes. J Mol Appl Genet. 1982;1(5):405–418. [PubMed] [Google Scholar]
- Schardl C. L., Kado C. I. A functional map of the nopaline catabolism genes on the Ti plasmid of Agrobacterium tumefaciens C58. Mol Gen Genet. 1983;191(1):10–16. doi: 10.1007/BF00330882. [DOI] [PubMed] [Google Scholar]
- Schrell A., Alt-Moerbe J., Lanz T., Schroeder J. Arginase of Agrobacterium Ti plasmid C58. DNA sequence, properties, and comparison with eucaryotic enzymes. Eur J Biochem. 1989 Oct 1;184(3):635–641. doi: 10.1111/j.1432-1033.1989.tb15060.x. [DOI] [PubMed] [Google Scholar]
- Sciaky D., Thomashow M. F. The sequence of the tms transcript 2 locus of the A. tumefaciens plasmid pTiA6 and characterization of the mutation in pTiA66 that is responsible for auxin attenuation. Nucleic Acids Res. 1984 Feb 10;12(3):1447–1461. doi: 10.1093/nar/12.3.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Zambryski P. C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986 Oct 24;47(2):155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- Vanderleyden J., Desair J., De Meirsman C., Michiels K., Van Gool A. P., Chilton M. D., Jen G. C. Nucleotide sequence of an insertion sequence (IS) element identified in the T-DNA region of a spontaneous variant of the Ti-plasmid pTiT37. Nucleic Acids Res. 1986 Aug 26;14(16):6699–6709. doi: 10.1093/nar/14.16.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B., Currier T. C., Gordon M. P., Chilton M. D., Nester E. W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975 Jul;123(1):255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Wheatcroft R. Nucleotide sequence of Rhizobium meliloti insertion sequence ISRm1: homology to IS2 from Escherichia coli and IS426 from Agrobacterium tumefaciens. DNA Seq. 1991;2(3):163–172. doi: 10.3109/10425179109039686. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Ebert P. R., Stachel S. E., Gordon M. P., Nester E. W. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8278–8282. doi: 10.1073/pnas.83.21.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]