Abstract
The Strongylocentrotus purpuratus sea urchin egg receptor for sperm is a cell surface glycoprotein with a molecular mass of 350 kDa. Recent studies indicate that the sulfated O-linked glycans isolated from the receptor bind to acrosome-reacted sperm. The purified receptor was analyzed with respect to amino acid and carbohydrate content and shown to be composed of 70% carbohydrate by weight. Compositional analysis indicated that both N- and O-linked oligosaccharide chains were present. After peptide:N-glycanase treatment of the receptor to remove most of the N-linked glycan chains, the majority of the sialic acid residues remained associated with the receptor and were shown by several types of experiments to be composed of sulfated oligosialic acid units attached to the O-linked glycan chains of the receptor. Chemical and physical studies on oligosialic chains discovered earlier in the Pronase-generated glycopeptide fraction isolated from the egg cell surface complex of another species of sea urchin, Hemicentrotus pulcherrimus, established that these molecules had the structure: (SO4−)-9Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n. Based on comparative and analytical studies, it was concluded that this sulfated oligosaccharide is a component of a GalNAc-containing chain that is O-linked to the polypeptide chain of the sea urchin egg receptor for sperm. Using a competitive inhibition of fertilization bioassay it was shown that the sulfated oligosialic acid chains derived from the S. purpuratus egg cell surface complex inhibited fertilization; the nonsulfated form of this oligosialic chain had little inhibitory activity.
Keywords: sulfated oligosaccharide, O-linked oligosaccharide, cell–cell interactions, fertilization
In fertilization, cell surface molecules of the egg and sperm play a central role in the species-specific interactions that occur in many organisms (1, 2). In the case of the sea urchin, earlier studies showed that the sperm protein, bindin, which is a component of the acrosome granule, plays a key role in gamete recognition (3, 4). Following early work in which a high molecular weight glycoprotein on the egg cell surface was implicated as a receptor for sperm (5–7), it was shown that Pronase-generated glycopeptides prepared from this crude receptor preparation inhibited fertilization, but without species specificity (8, 9). Later, a cell surface glycoprotein with a molecular mass of 350 kDa was identified as the egg receptor in Strongylocentrotus purpuratus (10, 11). This molecule, as well as a 70-kDa extracellular fragment generated by lysyl endoproteinase C digestion of intact eggs, were shown to contain sugars typical of both N- and O-linked oligosaccharides, as well as sulfate (10, 12). These oligosaccharide chains were fractionated, and the putative O-linked chains were shown to inhibit fertilization and bind without species specificity to acrosome-reacted, but not to unreacted sperm (13). In a subsequent study it was shown that attachment of these chains via a neoglycoprotein to beads mediated kinetically stable binding of sperm to such beads (14).
In earlier studies, α2→5-Oglycolyl-linked poly(Neu5Gc) (N-glycolylneuraminic acid) was identified for the first time in sialic acid (Sia)-rich glycoproteins (polySia-gp) purified from the egg jelly coat of two different species of sea urchins, Hemicentrotus pulcherrimus and S. purpuratus (15). In subsequent studies, sulfated oligosaccharide chains with the novel structure (SO4−)-9Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n were demonstrated to be present in the Pronase-generated glycopeptide fraction (ESP-Sia fraction) isolated from the egg cell surface complex (CSC) of the sea urchin, H. pulcherrimus (16). Preliminary experiments showed that this ESP-Sia fraction exhibited weak immunoreactivity against a polyclonal antibody that had been raised against the purified S. purpuratus egg receptor for sperm. These findings led us to investigate the possibility that sulfated oligosialic acid chains were a component of the 350-kDa S. purpuratus egg receptor for sperm. The results of these investigations revealed that, in fact, such sulfated oligosialic chains are O-linked via GalNAc residues to the polypeptide chain of egg receptor for sperm. It is of interest that earlier it was reported that glycoproteins containing both Neu5Gc and sulfate are present in the shell (test) of the sea urchin (17).
EXPERIMENTAL PROCEDURES
Materials.
Adult S. purpuratus sea urchins purchased from Marinus (Long Beach, CA) were maintained and gametes were collected as described (6). Peptide:N-glycanase (PNGase) was purchased from Boehringer Mannheim. The sea urchin preparations designated as the S. purpuratus polySia-gp, and H. pulcherrimus ESP-Sia fractions were prepared as described (15, 16).
Isolation of the 350-kDa Receptor Protein.
The recombinant protein designated 45A represents a major portion of the N-terminal domain of the egg receptor for sperm (11, 18). mAbs RS107 and RS132, which were prepared against the glutathione S-transferase–45A fusion protein (19) and protein A agarose were used for both immunoprecipitation and purification of the receptor. The S. purpuratus egg CSC consisting of plasma membrane, vitelline layer, and cortical granules was prepared as described (12).
For immunoprecipitation, a CSC containing 2 mg of protein was solubilized in 10 ml of 50 mM Tris·Cl (pH 8.0) containing 250 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 0.05% β-octyl glucoside, 0.5% Nonidet P-40, and 0.025% deoxycholate for 30 min on ice. After centrifugation at 3,000 × g for 15 min, the supernatant was incubated with the immuno beads at 4°C for 14 h. For subsequent analysis, the receptor was eluted and analyzed as described in Figs. 1 and 2.
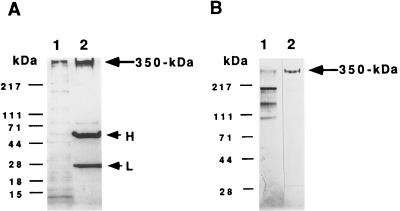
Figure 1.
Isolation of receptor by immunoaffinity chromatography. (A) Immunoprecipitation of S. purpuratus 350-kDa protein. In A, after incubation of the solubilized S. purpuratus egg cell CSC with mAb RS132 bound to protein A-agarose beads, the recovered beads were washed three times with TBS and then treated with SDS sample buffer containing 5% 2-mercaptoethanol and heated at 75°C for 15 min. Samples were then analyzed for bound proteins by 4–15% gradient SDS/PAGE under reducing conditions, followed by silver staining. Lanes: 1, solubilized egg CSC; 2, the proteins bound and eluted from the beads. H and L shows the positions of IgG heavy and light chains, respectively. (B) Analysis of receptor protein purified by immunoaffinity chromatography. For analysis, samples were electrophoresed on a 4–15% gradient SDS/polyacrylamide gel. Lanes: 1, solubilized egg CSC; 2, the fraction eluted with 5 M LiCl/10 mM Tris·Cl buffer (pH 7.4) from an affinity column made by covalently coupling mAb RS132 to protein A-agarose as described (20). Bio-Rad protein molecular markers: myosin (H-chain), 217 kDa; phosphorylase B, 111 kDa; BSA, 71 kDa; ovalbumin, 44 kDa; carbonic anhydrase, 28 kDa; β-lactoglobulin, 18 kDa; and lysozyme, 15 kDa.
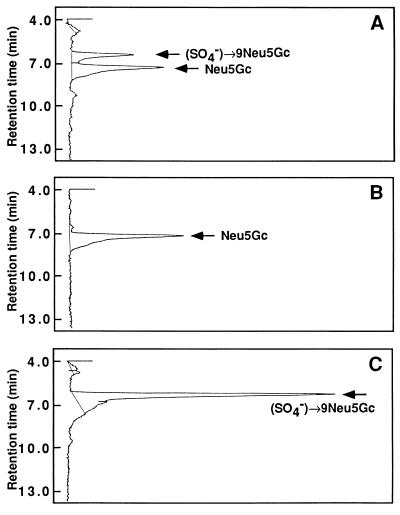
Figure 2.
Detection of sulfated Neu5Gc and Neu5Gc residues in receptor glycoprotein. The polyvinylidene difluoride membrane to which the 350-kDa protein was transferred from SDS/polyacrylamide gel was dipped in 1 ml of 0.1 M trifluoroacetic acid (TFA) and incubated in 80°C for 1 h. The solution containing the hydrolysate was transferred to a new tube and the acid was removed by evaporation. The residue was derivatized with 1,2-diamino-4,5 methyene dioxybenzene (Dojindo Laboratory, Kumamoto, Japan) for fluorometric HPLC (31) on a TSK-gel ODS-80 (150 × 4.6 mm interior diameter; particle size 5 μm; Tohso, Tokyo) using methanol/acetonitrile/0.05% (vol/vol) TFA (9:7:84, vol/vol) as the solvent system. TFA (0.015 M) was used as the acid during the derivatization procedure. (A) Elution profile for the mild acid hydrolysate of the receptor. (B and C) Elution profiles for Neu5Gc (15 pmol) and sulfated Neu5Gc (44 pmol) as reference, respectively. These reference compounds were isolated from rainbow trout polysialoglycoprotein (29) and sea urchin egg ESP-Sia (15), respectively.
The following (alternative) procedure was used to isolate a large amount of receptor in native form. Because the 350-kDa protein was found to be very sensitive to proteolytic degradation after homogenization of eggs, all the following procedures were carried out at 4°C and performed in the presence of protease inhibitors (2 μg/ml of soybean trypsin inhibitor/0.5 μg/ml Pefabloc SC/2 μg/ml of aprotinin/1 μg/ml of leupeptin/2 μg/ml of antipain/10 μg/ml of benzamidine/1 μg/ml of pepstatin/0.5 mM EDTA). The CSC (suspended in 5 ml of Ca2+-free sea water isolated from 2 ml of dejellied eggs) was solubilized using 2% (wt/vol) β-octyl glucoside (Boehringer Mannheim) as described (10). After insoluble material was removed by centrifugation at 10,000 × g for 20 min, the supernatant was diluted 1:4 (vol/vol) with Tris-buffered saline (TBS; 50 mM Tris·Cl, pH 7.4/150 mM NaCl) and applied to an immunoaffinity column, in which mAbs to the recombinant 45A protein (RS107 and RS132) were covalently coupled to protein A beads (20). After the column was washed with TBS buffer containing 0.1% (wt/vol) β-octyl glucoside, 5 M LiCl/10 mM Tris·Cl buffer (pH 7.4) was used to elute the 350-kDa protein. After extensive dialysis against distilled water, the receptor was stored at −20°C.
PNGase Treatment.
The purified 350-kDa protein (9.3 mg) was incubated with 14 milliunits of PNGase F in 3 ml of 50 mM Na-phosphate buffer (pH 7.4) containing 50 mM EDTA, 0.2% SDS, 1% Nonidet P-40, and 1% 2-mercaptoethanol for 36 hr at 37°C (21). Released N-glycans were separated from the de-N-glycosylated protein by performing Sephacryl S-200 gel filtration chromatography (1.4 × 35 cm).
β-Elimination of de-N-Glycosylated 350 kDa Protein.
The de-N-glycosylated 350 kDa protein (5.3 mg) was treated with 2 ml of 0.1 N NaOH-1M NaBH4 containing [3H]NaBH4 (25 mCi, 15 Ci/mmol; 1 Ci = 37 GBq; Amersham) at 37°C (16).
Preparation of Reference Oligosaccharides.
The (SO4−)-9Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n oligosaccharide (15) was prepared as follows: 500 ml of packed, dejellied S. purpuratus eggs were homogenized in Ca2+-free sea water containing protease inhibitors. The homogenate was centrifuged at 3,000 rpm in a refrigerated Sorvall bench-top centrifuge for 5 min. The pellets were resuspended and washed three times with cold Ca2+-free sea water. The final pellet, containing the egg CSC, was subjected to mild acid hydrolysis by incubation at 80°C for 12 hr in 50 mM Na-acetate buffer (pH 4.5). The mild acid hydrolysate was centrifuged at 3,000 rpm for 5 min, and ice-cold ethanol to a final concentration of 70% was added to the supernatant. After storage on ice for 15 min the preparation was centrifuged at 15,000 × g for 20 min. The supernatant was collected, concentrated, and desalted using a Sephadex G-25 column. The desalted sample was applied to a DEAE-Toyopearl 650 M column (2.2 × 10 cm) and eluted with 150 mM NaCl in 20 mM Tris·Cl buffer (pH 8.0).
The Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n and Neu5- Acα2(→8Neu5Acα2→)n oligosaccharides were prepared as described above by mild acid hydrolysis of S. purpuratus polySia-gp and colominic acid, respectively (16, 22). After desalting, these samples were loaded on a DEAE-Toyopearl column and eluted as described above. Based on monosaccharide analysis using a Dionex PA1 column, it was found that contamination with other oligosaccharide chains was less than 6%.
Determination of Interketosidic Linkage of Oligosialic Acid Chains.
Both the 350-kDa glycoprotein and, as a control, ESP-Sia containing (SO4−)-9Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n (20–50 μg as Sia) were subjected to strong alkali treatment and subsequent N-acetylation (15, 23, 24). The samples were desalted by passage through a Sephadex G-25 column and divided into two aliquots. One aliquot was treated with 2 milliunits of Arthrobacter ureafaciens sialidase (Oxford Glycosystems, Rosedale, NY) for 6 h at 37°C to release the glycoyl group. The second aliquot was incubated in the absence of sialidase.
Chemical Analysis.
Sia residues were quantified by the thiobarbituric acid method (25, 26), the resorcinol method, or by anionic-exchange HPLC on a Dionex PA1 column with pulsed amperometric detection (27). Sulfate ion analysis was performed after strong acid hydrolysis as described (16).
Amino sugar and neutral sugar analyses were also carried out on a Dionex PA1 column (28), and alditols were analyzed on a Dionex MA1 column (28). Amino acid analysis was carried out after hydrolysis in 6 N HCl at 105°C for 24 h under N2 (29). Protein was determined by the modified Lowry assay (BCA; Pierce).
Fertilization Assay.
Bioassays to study the effect of the purified oligosialic acid chain on fertilization were performed as described (8, 30) except that the eggs were suspended in 200 μl of artificial sea water containing 25 mM Hepes (pH 8.0).
RESULTS
Identification of Sulfated Neu5Gc and Neu5Gc Residues in the 350-kDa Receptor Glycoprotein.
As noted in the Introduction, preliminary experiments showed that ESP-Sia prepared from the cell surface fraction of H. pulcherrimus egg exhibited weak immunoreactivity against a polyclonal antibody that had been raised against the S. purpuratus egg receptor for sperm (data not shown). These results led us to investigate the possibility that the sulfated oligosialic chains found in this fraction might also be present as a component of the egg receptor for sperm. Initially, we undertook to determine if Sia and sulfated Sia could be detected in the isolated S. purpuratus egg receptor for sperm. The receptor was immunoprecipitated from the S. purpuratus egg CSC using mAb RS132. Part of the immunoprecipitated sample was removed and analyzed by SDS/PAGE followed by staining with silver. In Fig. 1A, lane 1 shows the solubilized egg CSC, and lane 2 shows the immunoprecipitated receptor and IgG. The remaining immunoprecipitated material was suspended in Laemmli sample buffer, subjected to preparative SDS/PAGE, and transblotted onto a polyvinylidene difluoride membrane. The region of the membrane corresponding to the mobility of the 350-kDa receptor was excised and analyzed for sulfated Neu5Gc and Neu5Gc residues following mild acid hydrolysis. The hydrolysates were transferred to a microtube, evaporated to dryness, and subjected to fluorometric HPLC analysis after derivatization. As shown in Fig. 2, sulfated Neu5Gc and Neu5Gc residues were readily be identified in the hydrolysate. The amounts of sulfated Neu5Gc and Neu5Gc were quantitated by comparing the peak area of each peak with those of authentic samples shown in Fig. 2 B and C.
Purification and Carbohydrate Analysis of the Receptor Protein.
For further analysis, we scaled up the immunopurification procedure and used a mAb (RS132) to recombinant 45A that was covalently coupled to protein A. The results of Fig. 1B shows SDS/PAGE analysis of the solubilized CSC (lane 1) and the purified 350-kDa protein recovered from the mAb RS132-protein A agarose column (lane 2). Initially, we used only mAb RS132 for purification of the 350-kDa protein. Later, we used a two-step procedure involving binding to mAb RS132, followed by binding of the eluant to a second antibody (mAb RS107) directed against the recombinant 45A protein. This mAb when bound to protein A agarose had a 2-fold greater binding capacity for the receptor protein than mAb RS132-protein A agarose. Using this procedure, we purified 3.6 mg of 350-kDa protein from 35 mg of total protein in a CSC preparation isolated from 2 ml of packed dejellied eggs. We sometimes observed immunoreactive proteins of lower molecular weight at 220, 190, 180, 140, and 105 kDa in the sample eluted from the immunoaffinity column. It is well known that sea urchin eggs contain a trypsin-like protease activity (32). However, in spite of the addition of protease inhibitors, we could not totally block degradation of the receptor protein. Because these lower molecular proteins were found to be recognized by several other antireceptor antibodies, it is clear that they are degradation products of 350-kDa protein (data not shown).
The receptor protein contained Sia that could be detected by means of the resorcinol method, the thiobarbituric acid method, and by anion-exchange HPLC on Dionex with pulsed amperometric detection. A sample containing 1 mg of the receptor protein determined by BCA assay contained 87 μg of Sia determined by resorcinol method. Analysis of the S. purpuratus receptor protein is shown in Table 1, column 2. The amount of carbohydrate associated with the 350-kDa protein was found to be 70% (wt/wt), which is consistent with that found in earlier studies. From this value it could be calculated that the polypeptide chain makes up 105 kDa of the total mass of the 350-kDa glycoprotein. This value is in good agreement with the 99 kDa calculated for the recently revised deduced amino acid sequence of the protein (18). Other components we detected were in accord with previous observations (13), namely Fuc, GalNAc, GlcNAc, Gal, and Man, which is consistent with the presence of both N- and O-linked oligosaccharides. Sulfate was also a component of the receptor. The ratio of total Neu5Gc to sulfate was determined to be 2.8:1. Quantitation of sulfated Neu5Gc and Neu5Gc residues in the receptor protein by means of fluorometric HPLC analysis indicated that the ratio of sulfated Neu5Gc to Neu5Gc was 1:1.2—i.e., the ratio of total Neu5Gc to sulfate was 2.2:1. Taken together, we can conclude that sulfate group attached to Neu5Gc accounted for the majority of the sulfate groups detected in the receptor.
Table 1.
Carbohydrate composition of the receptor glycoprotein after various treatments
| Carbohydrate composition after:
|
|||
|---|---|---|---|
| No treatment | PNGase treatment* | PNGase treatment followed by β-elimination† | |
| Amino acids (30%, wt/wt) | |||
| Carbohydrate (70%, wt/wt) | |||
| GalNAcol | 0 | 0 | 1.0 |
| Fuc | 0.5 | 0.1 | 0.1 |
| GalNAc | 1.6 | 1.6 | 0.3 |
| GlcNAc | 1.2 | 0.3 | 0.2 |
| Gal | 0.8 | 0.7 | 0.1 |
| Man | 13 | <0.2 | <0.2 |
| Neu5Gc | 3.8‡ (3.8§) | 3.8‡ | 3.8‡ |
| Sulfate | 1.3 (1.7§) | NA | NA |
NA, not analyzed.
After PNGase treatment released N-glycans were removed by Sephacryl S-200 gel filtration chromatography. One aliquot of the de-N-glycosylated receptor recovered in the void volume was used for carbohydrate compositional analysis and a second was used to carry out β-elimination.
The de-N-glycosylated sample was treated with NaOH-[3H]NaBH4 as described in Experimental Procedures. The reaction mixture was desalted on Sephadex G-25 and the labeled oligosaccharide recovered was used for carbohydrate analysis.
Molar ratios are relative to Neu5Gc taken as 3.8 setting the value for GalNAcol as 1.0.
The ratio was independently determined by quantitating sulfated Neu5Gc and Neu5Gc residues.
Evidence for the Presence of Sulfated oligoNeu5Gc Residues in the Receptor Glycoprotein and Determination of the Interresidue Linkage of the oligoNeu5Gc.
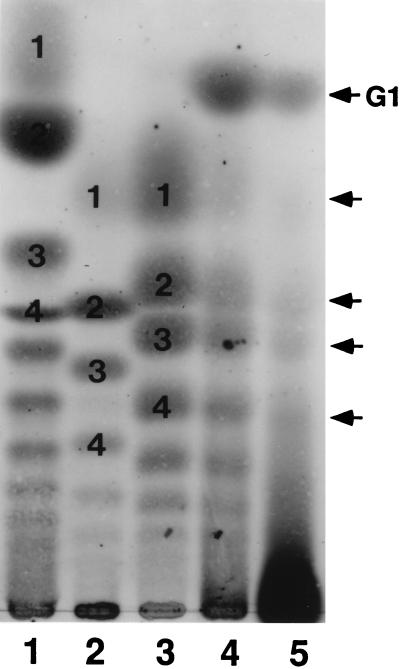
The compositional analysis presented in Table 1 showed that the majority of the O-glycan chains on the receptor glycoprotein contain one GalNAc unit presumably attached to a Ser/Thr residue of the core protein and about 4 (on average) Neu5Gc units. The low level of other sugar components suggested that the most, if not all, Neu5Gc residues are present in the form of oligoSia chains directly attached to the proximal GalNAc residue. To demonstrate directly the occurrence of oligoNeu5Gc chains and to obtain information on the type of interresidue linkage of Neu5Gc, we performed TLC analysis of the mild acid hydrolysate of the receptor under the previously established conditions (16, 33). We utilized hydrolytic conditions (pH 4.5, 80°C) under which the inter-residue linkage of unsulfated α2→5-Oglycolyl-linked oligo/polyNeu5Gc chains are known to be stable (16, 33), but a series of sulfated α2→Oglycolyl-linked (Neu5Gc)n (n = 1–7) are cleaved from ESP-Sia isolated from the CSC of H. pulcherrimus. Following such mild acid hydrolysis of the receptor, a series of sialyloligomers visualized by the resorcinol reagent was detectable on TLC. The mobilities of these sialyloligomers and relative intensities of the spots coincided with those from the products from ESP-Sia (Fig. 3, lane 5 vs. lane 4), but not with those for the products from α2→8-linked (Neu5Ac)n (lane 1) or (Neu5Gc)n (lane 2) chains. It should be pointed out that a series of sialyloligomers were also generated from the unsulfated α2→5-Oglycolyl-linked oligo/polyNeu5Gc chains present in polySia-gp isolated from the egg jelly of S. purpuratus, and the mobilities of these sialyloligomers appeared to be identical to those derived from ESP-Sia and the receptor glycoprotein. However, the more stringent mild acid hydrolysis conditions (0.1 M TFA, 65°C, 20 min) must be applied for this polymer to generate sialyloligomers (lane 3). The most important observation is the detection of a component, which migrated faster than the Neu5Gc monomer, in the mild acid hydrolysate of the receptor (lane 5). On the basis of its mobility, this component can be identified as (SO4−)-Neu5Gc (lane 4), whose structure was established previously (16). The distribution of the sialyloligomers in the hydrolysate of the receptor indicated the presence of oligomeric chains up to DP-4, although the presence of higher oligomers cannot be excluded from the presence of unresolved slow moving components and relatively low yield of lower oligomers.
Figure 3.
TLC analysis of the products of mild acid hydrolysis of the receptor. Approximately 40 μg as Sia was spotted on a silica gel plate, developed with 1-propanol/25% ammonia/water (6:1:2.5) for 12 h, and visualized by the resorcinol reagent (22). The following polySia-related molecules were hydrolyzed under the conditions indicated in parenthesis. Lanes: 1, colominic acid (pH 4.5, 80°C, 3 h); 2, rainbow trout polysialoglycoprotein (pH 4.5, 80°C, 3 h); 3, sea urchin S. purpuratus polySia-gp (0.1 M TFA, 65°C, 20 min); 4, H. pulcherrimus ESP-Sia (pH 4.5, 80°C, 5 h); 5, 350-kDa receptor protein (pH 4.5, 80°C, 15 h). The dark spot at the origin is not a resorcinol positive substance since it is brown, rather than purple. Numbers in lanes 1-3 represent the degree of polymerization. G1 in lane 5 is (SO4−)-9Neu5Gc. The arrowheads indicate the eluted position of NeuGc monomer (Upper) and α2→5-Oglycolyl-linked Neu5Gc di, tri, and tetramer (Lower).
The receptor was treated with strong alkali under conditions previously employed to establish that the interketosidic linkage in sea urchin egg jelly polySia-gp and the ESP-Sia fraction contained the α2–5→Oglycolyl (15, 16, 23) linkage. The TLC mobility of the product, visualized by the resorcinol reagent, corresponded with that of Neu5Acα2→OCH2COOH (data not shown). No other resorcinol-positive products were detected. Following treatment with a A. ureafaciens sialidase treatment the reaction product was converted into Neu5Ac monomer, which was identified by TLC. These results indicate that the interketosidic bonds in the sulfated oligoSia chain in the 350-kDa protein were α2→5-Oglycolyl linkages.
Presence of Sulfated oligoSia Chains in the O-Linked Glycans of the Receptor.
To determine whether sulfated oligoSia chains are attached to the O- or N-linked glycan chains in the 350-kDa glycoprotein, we undertook to remove N-linked glycans by PNGase F treatment to analyze only the remaining O-linked glycans. Although it was previously reported (13) that PNGase F did not act on the 350-kDa glycoprotein judging from a lack of change in its mobility upon SDS/PAGE, the more direct results of carbohydrate compositional analysis showed that PNGase F did cleave chains from the protein (Table 1, columns 2 and 3). After PNGase F treatment, the amount of Man residue was dramatically decreased suggesting most of the N-linked glycans were removed. Because after PNGase F treatment the Sia residues remained associated with the protein, we assumed that they were components of the O-linked glycans.
In view of these findings, we carried out β-elimination under reducing conditions using [3H]NaBH4 to release the O-linked glycans. The reduced sample was desalted on Sephadex G-25 and then fractionated on Sephadex G-50 to separate the liberated O-glycan from the peptide fraction. Most of the [3H]-labeled radioactivity was eluted after the void volume in a broad peak with apparent mass range of 1,700–600 Da. These results suggested that the major core O-linked oligosaccharide alditol is relatively short, in agreement with the results in Table 1, column 4, in which the major sugars detected were Neu5Gc and GalNAcol. The elution pattern of radioactivity was very similar when the β-eliminated ESP-Sia was fractionated by gel filtration and then analyzed for Sia.
The main labeled gel filtration peak was desalted on Sephadex G-25 column, and then fractionated on a DEAE-Toyopearl 650 M column that was eluted stepwise using a NaCl-containing buffer. The relative amount of β-eliminated, [3H]O-glycan in each fraction was calculated by quantitating the amount of [3H]GalNAcol after strong acid hydrolysis followed by Dionex MA-1 column analysis (Table 2). As shown, 15% of the labeled O-glycans were uncharged; ≈80% of the remaining O-glycans was eluted by either 50 mM or 100 mM NaCl-containing buffer. Less than 6% of O-glycans were eluted by 200 mM and 500 mM NaCl-containing buffer (data not shown). The average degree of polymerization of the sulfated oligoSia chains was relatively low, with a value of 3.8, if it is assumed that all Sia residues exist in linear sulfated oligoSia chains. It is noteworthy that the separation of the O-linked glycans by anionic exchange chromatography seemed to be based on the number of Sia residues. A (SO4−)-Neu5Gc residue is known to be present as the nonreducing terminal residue of oligo Neu5Gcα2 (→5-OglycolylNeu5Gcα2→)n chains in the H. pulcherrimus ESP-Sia fraction (15).
Table 2.
Characterization of the [3H]-labeled oligosaccharide alditols liberated from the receptor glycoprotein after PNGase treatment
| Total O-glycans | FT | 50 mM NaCl | 100 mM NaCl | |
|---|---|---|---|---|
| % of total O-glycans* | 100 | 15 | 53 | 26 |
| GalNAcol | 1.0† | 1.0† | 1.0† | 1.0† |
| Neu5Gc | 3.0 | 0 | 2.0 | 3.7 |
| Sulfated Neu5Gc | 0.5 | 0 | 0.3 | 0.7 |
After PNGase treatment, the glycoprotein was treated with NaOH-[3H]NaBH4, as described in Experimental Procedures. The released mixture of oligosaccharide alditols were subjected to anion exchange chromatography, and the flow-through fraction (FT) and the fractions eluted with 50 mM NaCl/10 mM Tris·Cl (pH 8.0) and then with 100 mM NaCl/10 mM Tris·Cl (pH 8.0) were analyzed.
The amount of O-glycan in each fraction was determined by quantitating the amount of [3H]GalNAcol after acid hydrolysis using a Dionex MA-1 column.
Molar ratios are relative to GalNAcol taken as 1.0.
Fertilization Inhibitory Activity of Sulfated oligoSia Chains.
In earlier studies, sulfated O-linked oligosaccharide chains were shown to have higher activity than the unsulfated O-linked chains in inhibiting fertilization in the competitive bioassay (13). In view of the finding that the sulfate is linked to oligoSia units attached to the O-linked chains, we tested the inhibitory activity of the sulfated oligoSia chains isolated from the egg CSC. As a control, we also tested the nonsulfated oligoSia chains and α2→8-linked (Neu5Ac)n oligosaccharide chains that were prepared by mild acid hydrolysis of the polySia containing glycoprotein of sea urchin egg jelly polySia-gp and colominic acid, respectively. All three oligoSia chains had roughly the same average degree of polymerization (≈4) as judged from TLC. As compared with the other oligoSia chains, only the (SO4−)-9Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n chain had inhibitory activity in the fertilization bioassay in the 600 μM range (Table 3).
Table 3.
Inhibitory activity of several types of oligoSia chains in fertilization assays
| Compound tested | % fertilization |
|---|---|
| (SO4−)9-Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n | 26 ± 11 |
| Neu5Gcα2(→5-OglycolylNeu5Gcα2→)n | 83 ± 21 |
| Neu5Acα2(→8Neu5Acα2→)n | 81 ± 7 |
Each value is the mean ± SE of three separate experiments. In all cases 600 μM oligosaccharide was added.
DISCUSSION
A cell surface glycoprotein with an apparent mass of 350 kDa has been identified as the sea urchin egg receptor for sperm in S. purpuratus (10). Early studies implicated not only the polypeptide backbone of the receptor, but the carbohydrate chains as well, in the sperm binding process (8, 9). More recently it has been shown that sulfated O-linked glycan chains isolated from the receptor by hydrazinolysis competitively inhibit fertilization in a competition bioassay (13). In addition, it was shown that the O-linked oligosaccharide chains labeled at their reducing terminus by reduction with [3H]NaBH4, bound to acrosome-reacted, but not unreacted sperm. Both the inhibition and the binding processes occurred without species specificity, as expected based on earlier studies (2, 7–9). More recently, it has been shown that coupling these chains to proteins produced neoglycoproteins that inhibited fertilization; the inhibition was proportional to the valency of the oligosaccharide in the neoglycoprotein (14). Further, it was found that these neoglycoproteins, when coupled to beads, increased the stability of binding of sperm to the beads (14).
Given these observations on the potential role of oligosaccharide chains in gamete binding, it was important to understand the structure of these chains. To accomplish this the receptor was isolated by immunoaffinity chromatography using a mAb against a recombinant protein representing a part of the N-terminal domain of the receptor. The resulting 350-kDa glycoprotein was subjected to a variety of chemical procedures that established the presence of sulfated oligoNeu5Gc chains that are attached to O-linked oligosaccharides on the sperm receptor. Further studies revealed that each Neu5Gc residue was linked to the hydroxyl group of the glycolyl moiety of the penultimate Neu5Gc residue. This conclusion was based on comparison of the polysialic acid chain in the receptor with the unique type of polySia, α2→5-Oglycolyl-polyNeu5Gc, that was first found in the jelly coat of sea urchin eggs (15). This novel structure was established in that system using NMR, fast atom bombardment-MS, and chemical analysis. The oligoSia chain presumably is linked at its reducing terminus to a truncated O-linked glycan of which the major component is a GalNAc residue. In the case of the receptor the oligoSia chain contains one sulfate unit per 2–4 Sia residues. Based on studies in H. pulcherrimus it seems likely that this sulfate is located at the nonreducing terminus of the chain, as part of the following structure: (SO4−)-9Neu5Gcα2-(→5-OglycolylNeu5Gcα2→)n.
We previously showed that the sulfated form of O-linked oligosaccharide chains of the receptor most potently inhibit fertilization (13). Our studies have now established that most, if not all, of the sulfate groups in the receptor protein are attached to Neu5Gc residues in the O-linked glycans and may be important in sperm binding. Interestingly, the sea urchin sperm adhesive protein, bindin, which binds to the egg surface molecule during fertilization, is known to specifically recognize sulfate esters on sulfated polysaccharides (34). This suggests that the sulfate residue in the receptor may function in the sperm binding process via interaction with bindin.
It is of interest to consider these findings in relation to the oligosaccharide chains of sperm binding molecules from other species. In the case of mouse, initial binding of sperm to eggs is believed to occur via O-linked oligosaccharide present on the ZP3 protein (35). Because either α-galactosidase or galactose oxidase treatment on ZP3 resulted in loss of sperm receptor activity, a galactose residue, located in α-linkage at the nonreducing terminus of O-linked oligosaccharides, is believed to be at least one of the sugar determinants (36, 37). The nonreducing terminal GlcNAc residue in ZP3 is also believed to mediate binding of sperm because sperm surface β1,4-galactosyltransferase recognizes this structure (38). Although the essential role of the O-linked oligosaccharides in ZP3 seems clear, only limited information is available concerning their structure (36, 37), as compared with detailed information on the N-linked oligosaccharides (39, 40). Similarly, in the bivalve, unio elongatulus, O-linked oligosaccharides derived from a 220-kDa vitelline coat glycoprotein have been suggested to be responsible for sperm binding (41, 42) and although there is evidence that a fucose residue is involved in the binding properties of the 220-kDa glycoprotein, the structure of the oligosaccharide containing the Fuc has not been determined (41). Clearly, more detailed structural studies are essential before we will understand the precise role of the components of the oligosaccharide chains of sperm binding proteins in the gamete binding process.
Acknowledgments
We gratefully acknowledge Dr. Robert Haltiwanger (State University of New York, Stony Brook) for the use of the high pH anion exchange chromatograph with pulsed amperometric detection and thank Lorraine Conroy for preparation of this manuscript. This work was supported by National Institutes of Health Grants HD18590 and GM33184 to W.J.L. S.K.-K. was supported by a “Japanese Society for Promotion of Science” Postdoctoral Fellowship for research abroad.
ABBREVIATIONS
- CSC
cell surface complex
- Neu5Gc
N-glycolylneuraminic acid
- PNGase
peptide:N-glycanase
- Sia
sialic acid
- TFA
trifluoroacetic acid
- ESP-Sia fraction
Pronase-generated glycopeptide fraction
- polySia-gp
Sia-rich glycoprotein
References
- 1.Wassarman P M. Science. 1987;235:553–560. doi: 10.1126/science.3027891. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Bravo N, Lennarz W J. In: The Molecular Biology of Fertilization. Schatten H, Schatten G, editors. New York: Academic; 1989. pp. 21–36. [Google Scholar]
- 3.Trimmer J S, Vacquier V D. Annu Rev Cell Biol. 1986;2:1–26. doi: 10.1146/annurev.cb.02.110186.000245. [DOI] [PubMed] [Google Scholar]
- 4.Minor J E, Gao E H, Davidson E H. In: The Molecular Biology of Fertilization. Schatten H, Schatten G, editors. New York: Academic; 1989. pp. 773–778. [Google Scholar]
- 5.Tsuzuki H, Yoshida M, Onitake K, Aketa K. Biochem Biophys Res Commun. 1977;76:502–511. doi: 10.1016/0006-291x(77)90753-7. [DOI] [PubMed] [Google Scholar]
- 6.Schmell E, Earles B J, Breaux C, Lennarz W J. J Cell Biol. 1977;72:35–46. doi: 10.1083/jcb.72.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossignol D P, Roschelle A J, Lennarz W J. J Supramol Struct Cell Biochem. 1981;15:347–358. doi: 10.1002/jsscb.1981.380150405. [DOI] [PubMed] [Google Scholar]
- 8.Rossignol D P, Earls B J, Decker G L, Lennarz W J. Dev Bol. 1984;104:308–321. doi: 10.1016/0012-1606(84)90086-1. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Bravo N, Lennarz W J. Dev Biol. 1986;118:202–208. doi: 10.1016/0012-1606(86)90088-6. [DOI] [PubMed] [Google Scholar]
- 10.Ohlendieck K, Dhume S T, Partin J S, Lennarz W J. J Cell Biol. 1993;122:887–895. doi: 10.1083/jcb.122.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foltz K R, Partin J S, Lennarz W J. Science. 1993;259:1421–1425. doi: 10.1126/science.8383878. [DOI] [PubMed] [Google Scholar]
- 12.Foltz K R, Lennarz W J. J Cell Biol. 1990;111:2951–2959. doi: 10.1083/jcb.111.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhume S T, Lennarz W J. Glycobiology. 1995;5:11–17. doi: 10.1093/glycob/5.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Dhume S T, Stears R L, Lennarz W J. Glycobiology. 1996;6:59–64. doi: 10.1093/glycob/6.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Kitazume S, Kitajima K, Inoue S, Troy F A, II, Cho J-W, Lennarz W J, Inoue Y. J Biol Chem. 1994;269:22712–22718. [PubMed] [Google Scholar]
- 16.Kitazume S, Kitajima K, Inoue S, Haslam S M, Morris H R, Dell A, Lennarz W J, Inoue Y. J Biol Chem. 1996;271:6694–6701. doi: 10.1074/jbc.271.12.6694. [DOI] [PubMed] [Google Scholar]
- 17.Karamanos N K, Manouras A, Anagnostides S, Makatsori E, Tsegenidis T, Antonopoulos C A. Biochimie. 1996;78:171–182. doi: 10.1016/0300-9084(96)89502-2. [DOI] [PubMed] [Google Scholar]
- 18.Just, M. L. & Lennarz, W. J. (1997) Dev. Biol., in press.
- 19.Stears R. Ph.D. thesis. Stony Brook: State Univ. of New York; 1996. [Google Scholar]
- 20.Gersten D M, Marchalonis J J. J Immunol Methods. 1978;24:305–309. doi: 10.1016/0022-1759(78)90133-3. [DOI] [PubMed] [Google Scholar]
- 21.Norgard-Sumnicht K E, Roux L, Toomre D K, Manzi A, Freeze H H, Varki A. J Biol Chem. 1995;270:305–309. doi: 10.1074/jbc.270.46.27634. [DOI] [PubMed] [Google Scholar]
- 22.Kitazume S, Kitajima K, Inoue S, Inoue Y. Anal Biochem. 1992;202:25–34. doi: 10.1016/0003-2697(92)90200-q. [DOI] [PubMed] [Google Scholar]
- 23.Jennings H J, Roy R, Michon F. J Immunol. 1985;134:2651–2657. [PubMed] [Google Scholar]
- 24.Takasaki S, Mizuochi T, Kobata A. Methods Enzymol. 1982;83:263–268. doi: 10.1016/0076-6879(82)83019-x. [DOI] [PubMed] [Google Scholar]
- 25.Aminoff D. Biochem J. 1961;81:384–392. doi: 10.1042/bj0810384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida Y, Tsukada Y, Sugimori T. J Biochem (Tokyo) 1977;82:1425–1433. doi: 10.1093/oxfordjournals.jbchem.a131830. [DOI] [PubMed] [Google Scholar]
- 27.Olechno J D, Carter S R, Edwards W T, Gillen D G. Am Biotechnol Lab. 1987;5:38–50. [Google Scholar]
- 31.Hara S, Takemori Y, Yamaguchi M, Nakamura M, Ohkura Y. Anal Biochem. 1987;164:138–145. doi: 10.1016/0003-2697(87)90377-0. [DOI] [PubMed] [Google Scholar]
- 28.Hardy M R, Townsend R R, Lee Y C. Anal Biochem. 1988;170:54–62. doi: 10.1016/0003-2697(88)90089-9. [DOI] [PubMed] [Google Scholar]
- 29.Inoue S, Matsumura G. Carbohydrate Res. 1979;74:361–368. [Google Scholar]
- 30.Kinsey W H, Lennarz W J. J Cell Biol. 1981;91:325–331. doi: 10.1083/jcb.91.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glabe C G, Vacquier V D. J Cell Biol. 1977;75:410–421. doi: 10.1083/jcb.75.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitazume S, Kitajima K, Inoue S, Troy F A, II, Lennarz W J, Inoue Y. Biochem Biophys Res Commun. 1994;205:893–898. doi: 10.1006/bbrc.1994.2748. [DOI] [PubMed] [Google Scholar]
- 34.DeAngelis P L, Glabe C G. Biochim Biophys Acta. 1990;1037:100–105. doi: 10.1016/0167-4838(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 35.Florman H M, Wassarman P M. Cell. 1985;41:312–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bleil J D, Wassarman P M. Proc Natl Acad Sci USA. 1988;85:6778–6782. doi: 10.1073/pnas.85.18.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Litscher E S, Juntunen K, Seppo A, Penttila L, Niemela R, Renkone O, Wassarman P M. Biochemistry. 1995;34:4662–4669. doi: 10.1021/bi00014a020. [DOI] [PubMed] [Google Scholar]
- 38.Miller D J, Macek M B, Shur B D. Nature (London) 1992;357:589–593. doi: 10.1038/357589a0. [DOI] [PubMed] [Google Scholar]
- 39.Mori E, Takasaki S, Hedrick J L, Wardrip N J, Kobata A. Biochemistry. 1991;30:2078–2087. doi: 10.1021/bi00222a012. [DOI] [PubMed] [Google Scholar]
- 40.Noguchi S, Nakano M. Biochim Biophys Acta. 1993;1158:217–226. doi: 10.1016/0304-4165(93)90018-4. [DOI] [PubMed] [Google Scholar]
- 41.Focarelli R, Rosati F. Dev Biol. 1995;171:606–614. doi: 10.1006/dbio.1995.1308. [DOI] [PubMed] [Google Scholar]
- 42.Rosati F. Boll Zool. 1995;62:323–334. [Google Scholar]