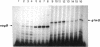
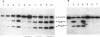Abstract
The intracellular concentration of the enzyme glucosamine-6-phosphate synthase, encoded by the gene glmS in Escherichia coli, is repressed about threefold by growth on the amino sugars glucosamine and N-acetylglucosamine. This regulation occurs at the level of glmS transcription. It is not due just to the presence of intracellular amino sugar phosphates, because mutations which derepress the genes of the nag regulon (coding for proteins involved in the uptake and metabolism of N-acetylglucosamine) also repress the expression of glmS in the absence of exogenous amino sugars.
Full text
PDF

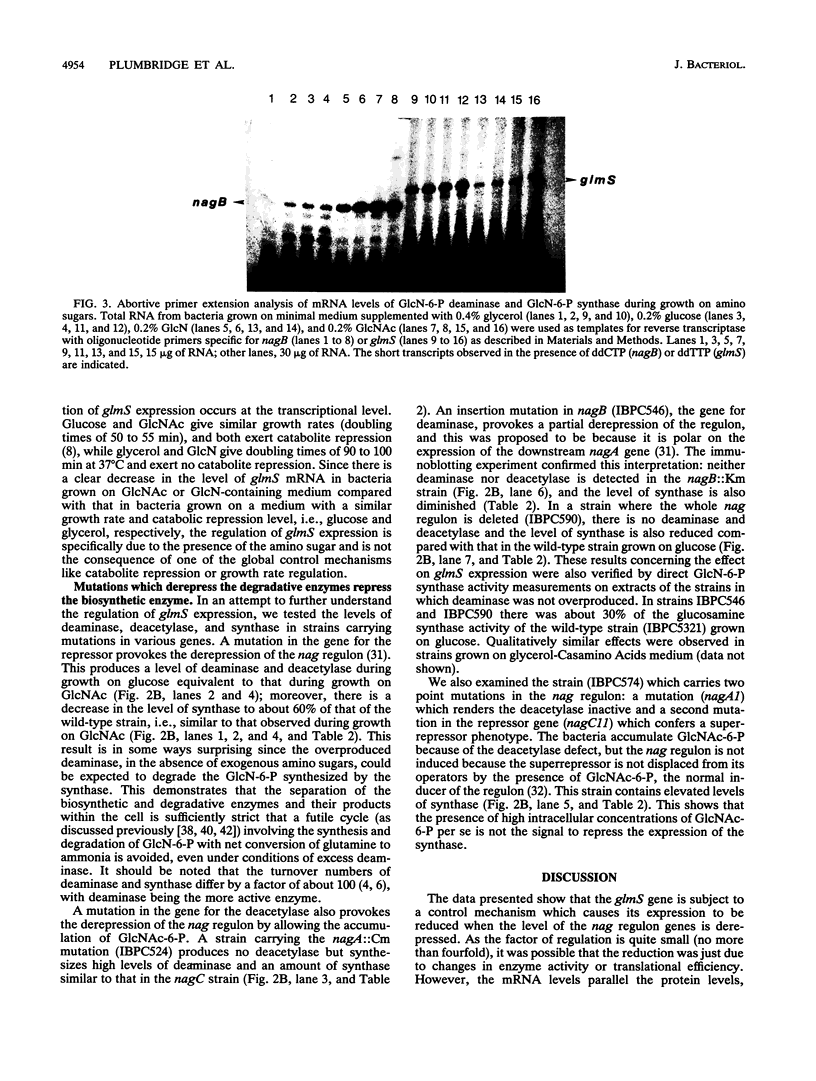


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamirano M. M., Plumbridge J. A., Hernández-Arana A., Calcagno M. Secondary structure of Escherichia coli glucosamine-6-phosphate deaminase from amino acid sequence and circular dichroism spectroscopy. Biochim Biophys Acta. 1991 Jan 29;1076(2):266–272. doi: 10.1016/0167-4838(91)90277-7. [DOI] [PubMed] [Google Scholar]
- Badet B., Vermoote P., Haumont P. Y., Lederer F., LeGoffic F. Glucosamine synthetase from Escherichia coli: purification, properties, and glutamine-utilizing site location. Biochemistry. 1987 Apr 7;26(7):1940–1948. doi: 10.1021/bi00381a023. [DOI] [PubMed] [Google Scholar]
- Badet B., Vermoote P., Le Goffic F. Glucosamine synthetase from Escherichia coli: kinetic mechanism and inhibition by N3-fumaroyl-L-2,3-diaminopropionic derivatives. Biochemistry. 1988 Apr 5;27(7):2282–2287. doi: 10.1021/bi00407a006. [DOI] [PubMed] [Google Scholar]
- Calcagno M., Campos P. J., Mulliert G., Suástegui J. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from Escherichia coli. Biochim Biophys Acta. 1984 Jun 14;787(2):165–173. doi: 10.1016/0167-4838(84)90076-1. [DOI] [PubMed] [Google Scholar]
- Curtis S. J., Epstein W. Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol. 1975 Jun;122(3):1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrogosz W. J. Effect of amino sugars on catabolite repression in Escherichia coli. J Bacteriol. 1968 Feb;95(2):578–584. doi: 10.1128/jb.95.2.578-584.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrogosz W. J. N-acetylglucosamine assimilation in Escherichia coli and its relation to catabolite repression. J Bacteriol. 1968 Feb;95(2):585–591. doi: 10.1128/jb.95.2.585-591.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisa P. S., Sonneborn D. R. Developmentally regulated interconversions between end product-inhibitable and noninhibitable forms of a first pathway-specific enzyme activity can be mimicked in vitro by protein dephosphorylation-phosphorylation reactions. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6289–6293. doi: 10.1073/pnas.79.20.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerse R. H., van der Pluijm J., Postma P. W. The repressor of the PEP:fructose phosphotransferase system is required for the transcription of the pps gene of Escherichia coli. Mol Gen Genet. 1989 Aug;218(2):348–352. doi: 10.1007/BF00331288. [DOI] [PubMed] [Google Scholar]
- Horjales E., Altamirano M. M., Calcagno M. L., Dauter Z., Wilson K., Garratt R. C., Oliva G. Crystallization and preliminary crystallographic studies of glucosamine-6-phosphate deaminase from Escherichia coli K12. J Mol Biol. 1992 Aug 20;226(4):1283–1286. doi: 10.1016/0022-2836(92)91068-z. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B. Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine 1-phosphate uridyltransferase in Bacillus subtilis. J Bacteriol. 1992 Nov;174(21):6852–6856. doi: 10.1128/jb.174.21.6852-6856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mortimer M. C., Kornberg H. L. Amino-sugar transport systems of Escherichia coli K12. J Gen Microbiol. 1980 Apr;117(2):369–376. doi: 10.1099/00221287-117-2-369. [DOI] [PubMed] [Google Scholar]
- Kornfeld R. Studies on L-glutamine D-fructose 6-phosphate amidotransferase. I. Feedback inhibition by uridine diphosphate-N-acetylglucosamine. J Biol Chem. 1967 Jul 10;242(13):3135–3141. [PubMed] [Google Scholar]
- Kucharczyk N., Denisot M. A., Le Goffic F., Badet B. Glucosamine-6-phosphate synthase from Escherichia coli: determination of the mechanism of inactivation by N3-fumaroyl-L-2,3-diaminopropionic derivatives. Biochemistry. 1990 Apr 17;29(15):3668–3676. doi: 10.1021/bi00467a012. [DOI] [PubMed] [Google Scholar]
- McKnight G. L., Mudri S. L., Mathewes S. L., Traxinger R. R., Marshall S., Sheppard P. O., O'Hara P. J. Molecular cloning, cDNA sequence, and bacterial expression of human glutamine:fructose-6-phosphate amidotransferase. J Biol Chem. 1992 Dec 15;267(35):25208–25212. [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982 Sep;151(3):1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983 Jun;154(3):1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri K. G., Goldie H., Waygood E. B. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem Cell Biol. 1990 Jan;68(1):123–137. doi: 10.1139/o90-017. [DOI] [PubMed] [Google Scholar]
- Pittard A. J., Davidson B. E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991 Jul;5(7):1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A. A dominant mutation in the gene for the Nag repressor of Escherichia coli that renders the nag regulon uninducible. J Gen Microbiol. 1992 May;138(5):1011–1017. doi: 10.1099/00221287-138-5-1011. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J Bacteriol. 1990 May;172(5):2728–2735. doi: 10.1128/jb.172.5.2728-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991 Aug;5(8):2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol Microbiol. 1989 Apr;3(4):505–515. doi: 10.1111/j.1365-2958.1989.tb00197.x. [DOI] [PubMed] [Google Scholar]
- Rogers M. J., Ohgi T., Plumbridge J., Söll D. Nucleotide sequences of the Escherichia coli nagE and nagB genes: the structural genes for the N-acetylglucosamine transport protein of the bacterial phosphoenolpyruvate: sugar phosphotransferase system and for glucosamine-6-phosphate deaminase. Gene. 1988;62(2):197–207. doi: 10.1016/0378-1119(88)90558-6. [DOI] [PubMed] [Google Scholar]
- Uzan M., Favre R., Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler A. P., Lengeler J. W. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol Gen Genet. 1989 Oct;219(1-2):97–105. doi: 10.1007/BF00261163. [DOI] [PubMed] [Google Scholar]
- Vogler A. P., Trentmann S., Lengeler J. W. Alternative route for biosynthesis of amino sugars in Escherichia coli K-12 mutants by means of a catabolic isomerase. J Bacteriol. 1989 Dec;171(12):6586–6592. doi: 10.1128/jb.171.12.6586-6592.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Gay N. J., Saraste M., Eberle A. N. DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J. 1984 Dec 15;224(3):799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968 Feb;106(4):847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn P. J., Phelps C. F. Studies on the control of hexosamine biosynthesis by glucosamine synthetase. Biochem J. 1971 Feb;121(4):711–720. doi: 10.1042/bj1210711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C., Wu T. C. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J Bacteriol. 1971 Feb;105(2):455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]