Abstract
The E3 ubiquitin-protein ligases play an important role in controlling substrate specificity of the ubiquitin proteolysis system. A biochemical approach was taken to identify substrates of Rsp5, an essential hect (homologous to E6-AP carboxyl terminus) E3 of Saccharomyces cerevisiae. We show here that Rsp5 binds and ubiquitinates the largest subunit of RNA polymerase II (Rpb1) in vitro. Stable complex formation between Rsp5 and Rpb1 was also detected in yeast cell extracts, and repression of RSP5 expression in vivo led to an elevated steady-state level of Rpb1. The amino-terminal domain of Rsp5 mediates binding to Rpb1, while the carboxyl-terminal domain of Rpb1, containing the heptapeptide repeats characteristic of polymerase II, is necessary and sufficient for binding to Rsp5. Fusion of the Rpb1 carboxyl-terminal domain to another protein also causes that protein to be ubiquitinated by Rsp5. These findings indicate that Rsp5 targets at least a subset of cellular Rpb1 molecules for ubiquitin-dependent degradation and may therefore play a role in regulating polymerase II activities. In addition, the results support a model for hect E3 function in which the amino-terminal domain mediates substrate binding, while the carboxyl-terminal hect domain catalyzes ubiquitination of bound substrates.
Keywords: RSP5, RPB1/carboxyl-terminal domain
The ubiquitin-dependent proteolysis system is characterized by the covalent ligation of multiple ubiquitin peptides to target proteins, which serves as a signal for recognition and degradation by the 26S proteasome (1, 2). Ubiquitination may in some cases serve a regulatory function independent of proteolysis (2). Three classes of enzymes are known to be involved in ligation of ubiquitin to protein substrates: the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzymes, and the E3 ubiquitin-protein ligases. The E3 proteins are thought to play the major role in determining the substrate specificity of the system. Characterization of the human papillomavirus E6-mediated degradation of p53 led to the discovery of the E6-AP ubiquitin-protein ligase, which forms a stable ternary complex with E6 and p53, then directly catalyzes the ubiquitination of p53 (3–6). Ubiquitination is dependent on a “thioester cascade,” in which ubiquitin is transferred from the active site cysteine of the E1 enzyme, to the active site cysteine of an E2 enzyme, then finally to the active site cysteine of E6-AP, which catalyzes isopeptide bond formation between ubiquitin and one or more lysine residues of the substrate.
The carboxyl-terminal domain of E6-AP [the hect domain (homologous to E6-AP carboxyl terminus)], consisting of ≈350 amino acids, is similar to that found in at least 30 other eukaryotic proteins (7). The active-site cysteine of E6-AP is within the hect domain and is absolutely conserved among all of the E6-AP-related proteins. Several of these proteins have been shown to also form ubiquitin-thioesters with the same requirements as E6-AP, strongly suggesting that these proteins represent a family of E3 ubiquitin-protein ligases. While other E3 proteins and activities have been identified (8–10), the hect E3s are so far the only known family of related E3 proteins.
The yeast Saccharomyces cerevisiae has five genes that encode hect E3 proteins, including the essential RSP5 gene, also known as NPI1 (7, 11). RSP5 was first identified in a search for suppressors of mutations in SPT3 (B. Berg, A. Happel, and F. Winston, cited in ref. 7), which encodes a protein that interacts with the TATA-binding protein (the SPT15 gene product) (12). We report here the results of a biochemical approach aimed at identifying substrates of Rsp5. This approach was based on our prior characterization of E6-AP, which suggested two criteria for identification of putative substrates: stable complex formation between the E3 and its substrates, and direct catalysis of substrate ubiquitination by the E3, with the latter being dependent on ubiquitin thioester formation at the active-site cysteine.
MATERIALS AND METHODS
Plasmids.
Generation of plasmids containing the RSP5 gene and the active-site Cys to Ala (C-A) mutant have been described (7). The full-length ORFs and fragments of RSP5 were subcloned into pYES2 (Invitrogen), pGEX-4T-1 (Pharmacia), or the pVL1393 baculovirus transfer vector. The RPB1 gene was amplified by PCR from yeast genomic DNA and cloned into pYES2. The carboxyl-terminal domain (CTD) of Rpb1 was amplified by PCR from this plasmid and subcloned into pGEX-4T-1. Escherichia coli expression vectors and the production of wheat E1, and Arabidopsis thaliana UBC8 and UBC1 has been described (5, 13, 14). The plasmid used in generating the GAL-RSP5 strain (see Fig. 4) was generated by subcloning nucleotides 1–1,077 of Rsp5 into pYES2, excising the 2-μm replication origin of pYES2 by digestion with NaeI and ClaI, then religating the plasmid. This plasmid was used to transform the FY56 yeast strain (MATα, his4-912δR5, lys2-128δ, ura3-52), selecting for growth on uracil drop-out plates with 2% galactose. Expression of glutathione S-transferase (GST) fusion proteins in yeast was performed by subcloning the GST ORF from pGEX4T-1 into pYES2.
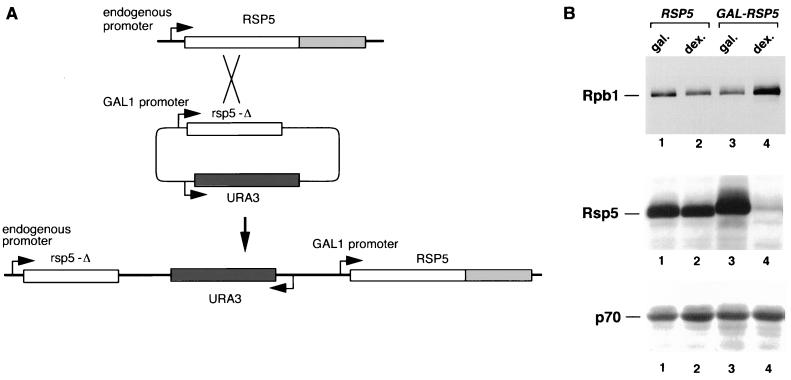
Figure 4.
(A) Generation of the GAL-RSP5 strain. Nucleotides 1–1,077 of the RSP5 gene (rsp5-Δ) were cloned into the pYES2 vector (Invitrogen) lacking a 2-μm replication origin. This was used to transform the FY56 yeast strain (ura3-52), selecting for growth on 2% galactose plates in the absence of uracil. The predicted recombination product, which was confirmed by PCR analysis of genomic DNA, is shown. (B) The RSP5 and GAL-RSP5 strains were grown at 30°C in minimal medium with 2% galactose to an OD600 of 0.5. Aliquots of the culture were removed to either fresh galactose- or dextrose-containing medium and incubated for an additional 36 h. Total cell extracts were analyzed by SDS/PAGE and immunoblotting with antibodies against Rpb1, Rsp5, and the 70-kDa subunit of yeast replication protein A (RPA). Extracts from the RSP5 strain, grown in galactose (Gal.) and dextrose (Dex.), respectively, were analyzed in lanes 1 and 2, and extracts from the GAL-RSP5 strain were analyzed in lanes 3 and 4. Rsp5 protein from the GAL-RSP5 strain migrates slightly slower due to a 12-amino acid epitope at its amino terminus.
Protein Expression.
In vitro translation reactions utilized pYES2 vectors and a coupled in vitro T7 transcription/translation system (TnT; Promega). GST fusion proteins were expressed in E. coli DH5α. Recombinant baculoviruses expressing Rsp5 or Rsp5 C-A proteins were generated using the BaculoGold system (PharMingen). Proteins were expressed by infection of insect cells (High5 cells; Invitrogen) and partially purified from extracts by batch chromatography on DEAE Sephacell (Pharmacia), as described previously for E6-AP (4).
35S-labeled yeast extracts (see Fig. 1) were prepared by labeling log-phase cultures of FY56 for 1 h in the presence of [35S]methionine and [35S]cysteine, then vortex mixing the cells with glass beads for 20 min in buffer containing 200 mM Tris (pH 8.0), 400 mM (NH4)2SO4, 1 mM EDTA, 10 mM MgCl2, 10% glycerol, 7 mM 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. The lysates were clarified by microcentrifugation at 4°C for 30 min. Unlabeled extracts were prepared by the same protocol (see Fig. 5). Yeast extracts for Western blot analyses (Fig. 4) were prepared by NaOH lysis followed by trichloroacetic acid precipitation (15).
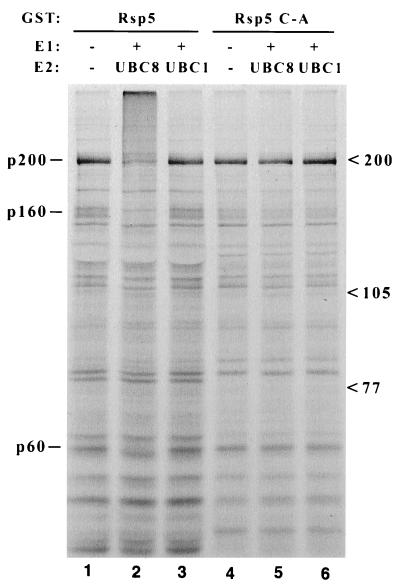
Figure 1.
Binding and ubiquitination of 35S-labeled yeast proteins by Rsp5. Glutathione-Sepharose-bound GST-Rsp5 fusion proteins, either wild type (lanes 1–3) or the C-A mutant (lanes 4–6), were incubated with 35S-labeled total yeast extract. The Sepharose beads were collected, washed, and then incubated without (lanes 1 and 4) or with (lanes 2, 3, 5, 6) recombinant E1 enzyme and UBC8 (lanes 2 and 5) or UBC1 (lanes 3 and 6) E2 protein. Proteins were then denatured in loading buffer and analyzed by SDS/PAGE and autoradiography. Three protein species that appear to be bound and ubiquitinated specifically by wild-type GST-Rsp5/UBC8 are indicated at left (p200, p160, p60), and migration positions of molecular weight markers are indicated at right.
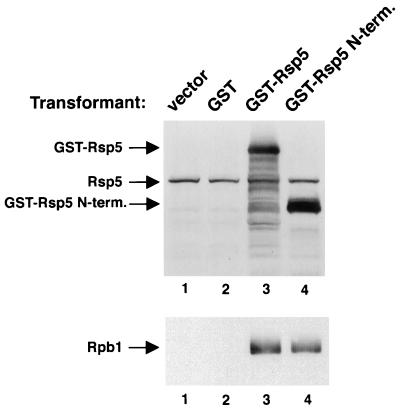
Figure 5.
The FY56 strain was transformed with the pYES2 vector or the vector expressing GST, GST-Rsp5, or GST-Rsp5 N terminus (lanes 1–4, respectively). Extracts were made from the cells and glutathione binding proteins, as well as copurifying proteins, were isolated on glutathione-Sepharose and analyzed by SDS/PAGE and immunoblotting. GST (nonfusion protein) expression was detected separately by silver staining of the glutathione-bound fraction (data not shown).
Protein Binding and Ubiquitination Assays and Protein Purification.
Binding reactions (as in Figs. 1, 2, and 5) contained 125 μl 25 mM Tris (pH 8.0), 125 mM NaCl, 0.1% Nonidet P-40, and 100 ng of GST fusion protein bound to 10 μl of glutathione-Sepharose. For binding reactions utilizing in vitro-translated proteins, 5 μl of programmed rabbit reticulocyte lysate was added to the reaction. Twenty-five microliters of 35S-labeled yeast extract (≈5 μg of total protein) was used in the binding reactions shown in Fig. 1. Reactions were rotated for 2 h at 4°C, then the Sepharose beads were washed three times with 500 μl of buffer containing 100 mM Tris, 100 mM NaCl, and 1% Nonidet P-40. SDS/PAGE loading buffer was added directly to the beads and heated at 95°C for 5 min; the released proteins were analyzed by SDS/PAGE and autoradiography.
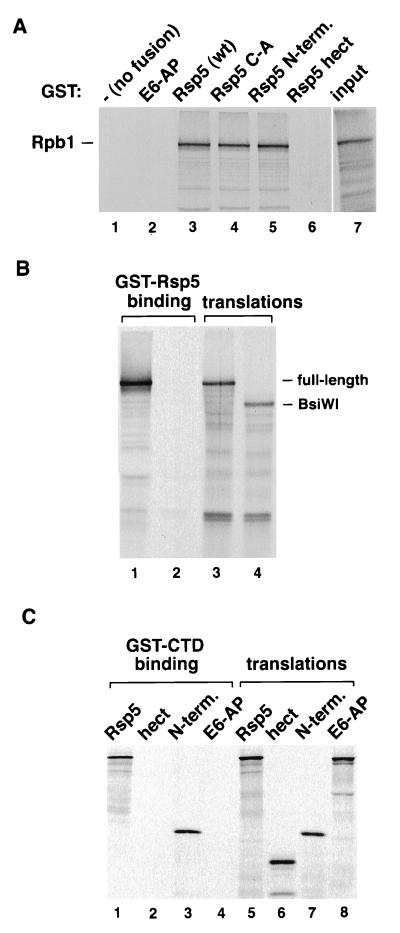
Figure 2.
(A) Binding of 35S-labeled in vitro-translated Rpb1 to GST (−, no fusion), or GST fused to E6-AP, Rsp5, the Rsp5 C-A mutant, the hect domain of Rsp5, or the N-terminal domain of Rsp5 (lanes 1–6, respectively). Five microliters of the translation reaction was used in the binding assays and was also loaded in lane 7. (B) Binding of carboxyl-terminally truncated Rpb1 to Rsp5. The Rpb1 gene in pYES2 was digested with BsiWI (nucleotide 4,528 of the 5,181 nucleotide ORF). The full-length and truncated genes were used in in vitro transcription/translation reactions. One microliter of the translation reactions was run in lanes 3 and 4. (C) Binding of Rsp5 to GST-CTD. In vitro-translated Rsp5, the Rsp5 hect domain, the Rsp5 amino-terminal domain (N-term), and E6-AP were assayed for binding to GST-CTD (lanes 1–4, respectively). One microliter of the translation products was run in lanes 5–8.
For ubiquitination of GST-Rsp5-bound proteins (Fig. 1), beads were incubated in a 75 μl volume containing 25 mM Tris (pH 8.0), 125 mM NaCl, 2 mM MgCl2, 50 μM DTT, 2 mM ATP, and 3 μg ubiquitin (Sigma), without or with E1 enzyme and UBC8 or UBC1. The reactions were incubated at room temperature for 1 h before addition of SDS/PAGE loading buffer and analyzed as described above. Ubiquitination assays using in vitro-translated Rpb1 or GST-CTD (Fig. 3) or crude HeLa extract (Fig. 6) were done under the same conditions.
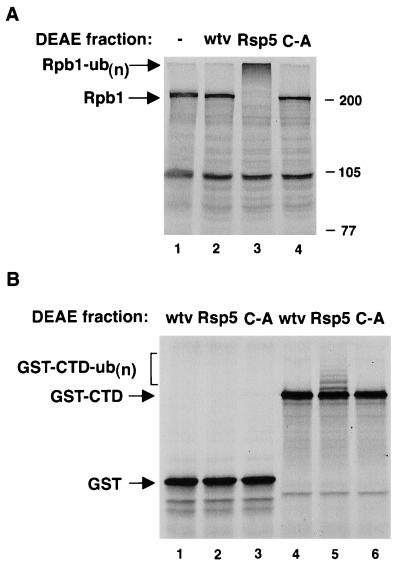
Figure 3.
(A) In vitro ubiquitination of Rpb1. In vitro-translated Rpb1 was incubated without (−, lane 1) or with a DEAE high-salt fraction derived from insect cells infected with nonrecombinant baculovirus (wtv, lane 2), Rsp5 virus (lane 3), or the C-A mutant (lane 4), along with recombinant E1 and E2 (UBC8) proteins, ATP, and ubiquitin. The migration position of full-length Rpb1 is indicated, as is multi-ubiquitinated Rpb1 [Rpb1-ub(n)]. (B) Ubiquitination of GST-CTD. In vitro-translated GST (lanes 1–3) or GST-CTD (lanes 4–6) were incubated with a control protein fraction (lanes 1 and 4), Rsp5 (lanes 2 and 5), or Rsp5 C-A (lanes 3 and 6), as in A. The ubiquitinated GST-CTD species are indicated.
Figure 6.
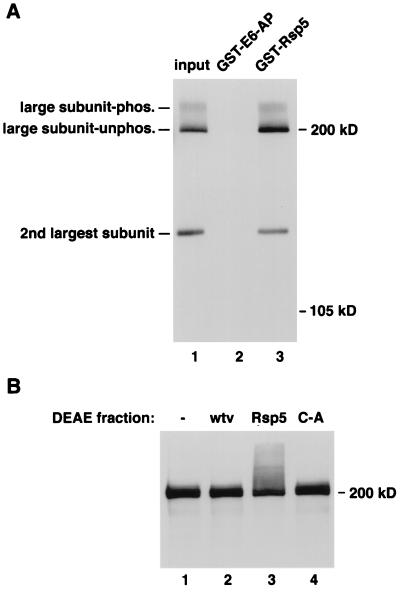
(A) Purified pol II from HeLa cells was assayed for binding to GST-E6-AP and Rsp5 (lanes 2 and 3). The largest and second largest subunits were detected by immunoblotting. The input amount of protein was loaded in lane 1. The phosphorylated and underphosphorylated forms of the largest subunit are indicated. (B) In vitro ubiquitination of human pol II large subunit. Total HeLa cell extract was incubated without (−, lane 1) or with a DEAE fraction from insect cells infected with nonrecombinant baculovirus (wtv, lane 2), Rsp5-expressing virus (Rsp5, lane 3), or Rsp5 C-A-expressing virus, along with ubiquitin, ATP, and E1 and UBC8 E2 proteins. The reactions were analyzed SDS/PAGE and immunoblotting with the anti-CTD antibody.
Binding reactions utilizing unlabeled proteins were analyzed by immunoblotting, using polyvinylidene fluoride (PVDF) membrane (Millipore) and detecting primary antibodies with horseradish peroxidase-linked secondary antibodies and the Renaissance chemiluminescent detection system (DuPont/NEN). Mouse mAbs were generated using purified GST-Rsp5 as antigen. Antibodies against the CTD and the second largest human polymerase II (pol II) subunit, as well as purified human pol II, were generously provided by Danny Reinberg and coworkers (Robert Wood Johnson University of Medicine and Dentistry of New Jersey). Antibodies against the 70-kDa subunit of replication protein A were provided by Steve Brill (Rutgers University).
Large-scale p200 isolation was done by preparing glass bead extracts from 10 liters of log-phase yeast culture and mixing this with 100 μg of GST-Rsp5 bound to 100 μl of glutathione-Sepharose. Mixing and washing conditions were identical to those described above for analytical binding assays. Following washing, the beads were heated in SDS/PAGE loading buffer and released proteins were loaded into a single lane of a 7% polyacrylamide gel. Proteins were then blotted to PVDF, stained with Ponceau S, and the band corresponding to p200 was cut from the membrane and sent to Harvard Microchemistry (Cambridge, MA) for analysis, peptide isolation, and sequencing. Two peptide sequences were obtained: EVQFGLFSPEEVRAISVAK and DQYSAPL. The first peptide corresponds exactly to residues 16–34 of Rpb1, while the second, which was of lower confidence, is similar but not identical to residues 4–11 of Rpb1 (QQYSSAPL).
Expression of GST fusion proteins in yeast was performed by growing the yeast transformants in synthetic minimal media with 2% dextrose to an OD600 of 1.0, then switching the cells to 2% galactose-containing media for 6 h. Extracts were made by glass bead vortex mixing and glutathione-binding proteins, along with copurifying proteins, were isolated on glutathione-Sepharose and analyzed by SDS/PAGE and immunoblotting.
RESULTS
35S-labeled yeast extracts were screened for proteins that bound to both wild-type GST-Rsp5 and GST-Rsp5 C-A, the active-site Cys to Ala mutant. The analogous C-A mutation in E6-AP does not affect its ability to bind to p53 but completely abrogates p53 ubiquitination (6). Putative substrates bound to wild-type Rsp5 were therefore predicted to be multi-ubiquitinated upon addition of the components necessary to active Rsp5 (ATP, ubiquitin, E1 protein, and an appropriate E2 protein), while they should not be ubiquitinated when bound to the catalytically inactive C-A mutant. Many proteins were observed to bind both GST-Rsp5 and GST-Rsp5 C-A, while a major protein species of ≈200 kDa (p200), and two less abundant species of ≈160 kDa and 60 kDa (p160 and p60), were detected which were both bound and ubiquitinated by wild-type GST-Rsp5 (Fig. 1). Ubiquitination was supported by the Arabidopsis thaliana UBC8 E2 protein, which is able to catalyze Rsp5-ubiquitin-thioester formation, but not by UBC1, which cannot activate Rsp5 (7).
Approximately 10 μg of p200 was isolated using GST-Rsp5 bound to glutathione-Sepharose as an affinity matrix. p200 was blotted from an SDS/polyacrylamide gel to PVDF membrane, and two peptides derived from this material were sequenced. Comparison against sequence databases indicated that p200 is the product of the RPB1 gene (calculated molecular mass, 191 kDa), the largest subunit of RNA pol II. p160 and p60 have not yet been characterized.
Rpb1 was expressed by in vitro translation and shown to bind to both GST-Rsp5 and the C-A mutant (Fig. 2A). In addition, Rpb1 bound to the amino-terminal domain of Rsp5 (amino acids 1–424), but not to the hect domain of Rsp5 (amino acids 460–809), or to GST or GST-E6-AP. These results indicate that the region of Rsp5 amino-terminal to the catalytic hect domain directs the interaction with Rpb1. Carboxyl-terminally deleted Rpb1 proteins were generated to map the region of Rpb1 necessary for recognition by Rsp5. A deletion of 217 amino acids (truncation at a unique BsiWI site) resulted in an inability to bind to Rsp5 (Fig. 2B). The BsiWI truncation eliminates the CTD of Rpb1, which contain 26 copies of a heptapeptide repeat characteristic of the largest subunit of all eukaryotic pol II molecules (16). To determine if the CTD is sufficient for binding to Rsp5, the carboxyl-terminal 205 amino acids of Rpb1 were assayed for binding to in vitro translated Rsp5 (Fig. 2C). GST-CTD bound to both full-length Rsp5 and the amino-terminal domain of Rsp5, but not to the Rsp5 hect domain or to E6-AP, indicating that the CTD of Rpb1 is both necessary and sufficient for stable association with Rsp5.
Baculovirus-expressed Rsp5 and Rsp5 C-A proteins were assayed for the ability to ubiquitinate Rpb1 in vitro. Rpb1 was incubated with either a control protein fraction or Rsp5 or Rsp5 C-A protein fractions, along with E1 and UBC8 E2 proteins (Fig. 3A). Rpb1 was multi-ubiquitinated only in the presence of wild-type Rsp5. While the ubiquitination was very efficient, the degradation of the ubiquitinated Rpb1 was variable in this system, probably due to low 26S proteasome activity in the reticulocyte lysate. Because the CTD was sufficient for Rsp5 binding, we also tested whether the CTD, when fused to another protein, was sufficient to direct Rsp5-dependent ubiquitination. As shown in Fig. 3B, incubation with wild-type Rsp5 led to the ubiquitination of in vitro-translated GST-CTD, although the ubiquitination was inefficient compared with that seen with Rpb1. GST by itself was not ubiquitinated by Rsp5. The lysine residue(s) of GST-CTD that are ubiquitinated by Rsp5 are likely to lie within the GST portion of the fusion protein and not within the CTD, since there are only two lysine residues within this region of Rpb1, which follow the last heptapeptide repeat, and these lysines can be deleted in the context of Rpb1 without affecting ubiquitination (not shown).
To determine if RSP5 affects Rpb1 steady-state levels in vivo, the RSP5 gene in a haploid yeast strain was disrupted and replaced with a GAL1 promoter-driven RSP5 gene (Fig. 4A). As expected, the GAL-RSP5 strain grows normally on galactose, but exhibits a growth defect on dextrose with a doubling time of ≈8 h, compared with about 1.5 h for the isogenic wild-type strain. Rsp5 and Rpb1 protein levels in the parental RSP5 strain did not differ between cells grown in galactose or dextrose. In contrast, after extended incubation of the GAL-RSP5 strain in dextrose-containing media (30–36 h), Rsp5 levels had decreased 30- to 50-fold, while Rpb1 levels increased ≈5-fold (Fig. 4B). The steady-state levels of a control protein, the 70-kDa subunit of replication protein A (17), did not differ between strains or carbon source. These results, consistent with the in vitro ubiquitination data, indicate that the steady-state level of Rpb1 is affected by Rsp5 in vivo.
To determine if Rsp5/Rpb1 complexes could be isolated from yeast cells, GST-Rsp5 and GST-Rsp5 N terminus were expressed in yeast. The GST-Rsp5 protein (wild type) was active in vivo since its expression could complement an rsp5 mutant (not shown). Lysates were made from the transformants 6 h after induction of fusion protein expression. Glutathione binding proteins, along with copurifying proteins, were isolated on glutathione-Sepharose and Rpb1 was assayed by immunoblotting (Fig. 5). Rpb1 copurified with GST-Rsp5 or GST-Rsp5 N terminus, but not with GST alone or from cells expressing no GST protein. By analyzing dilutions of the crude extract it was determined that ≈10% of the total Rpb1 was bound by GST-Rsp5.
Rsp5 was also shown to bind to the human homolog of Rpb1 in vitro (Fig. 6A). The pol II used in this experiment was an active, highly purified enzyme from HeLa cells. Because the phosphorylated and unphosphorylated forms of the largest subunit of human pol II are more easily resolved than with the yeast protein, this experiment also showed that Rsp5 can bind to both forms of the human protein. The same blot was probed with an antibody against the second largest subunit of human pol II. This subunit also complexed with GST-Rsp5 and Rpb1, presumably indirectly through its association with the largest subunit, indicating that Rsp5 can bind the largest subunit even when it is complexed with other core pol II subunits. Fig. 6B shows that incubation with baculovirus-expressed Rsp5 protein leads to the ubiquitination of the human Rpb1 homolog. While the phosphorylated and unphosphorylated forms were not clearly resolved in this experiment, additional experiments indicated that Rsp5 can ubiquitinate Rpb1 regardless of its phosphorylation state. The second largest subunit was not ubiquitinated when part of the Rsp5/large subunit complex (not shown).
DISCUSSION
Our results demonstrate that the largest subunit of yeast RNA pol II is a substrate of Rsp5. The ubiquitin system is generally associated with the turnover of rapidly degraded proteins; however, Rpb1 is a long-lived protein, with a half-life of at least 5 h. It therefore appears that Rsp5 does not target the majority of cellular Rpb1 for rapid degradation. The function of Rsp5 might be to control the steady-state concentration of Rpb1, which could be critical for either proper assembly of the pol II complex or for maintenance of a critical concentration of active pol II enzyme. Alternatively, since Rpb1 is present in many distinct complexes in the cell, Rsp5 might target a particular subpopulation of Rpb1 for rapid degradation. Our in vitro experiments have not suggested a particular Rpb1 subpopulation that might be targeted in vivo, since Rsp5 could target all of the Rpb1 complexes examined: uncomplexed Rpb1, Rpb1 in complex with other core pol II subunits, and both phosphorylated and unphosphorylated forms of Rpb1 (18, 19). The CTD is the apparent recognition site for several basal transcription factors and pol II holoenzyme components (20), as well as for Rsp5. It therefore seems likely that any specificity of Rsp5 for a particular pol II subpopulation might be based on CTD-associated transcriptional components.
It has recently been reported that the large subunit of human pol II is ubiquitinated following UV- or cisplatin-induced DNA damage (21). The ubiquitination was not observed in fibroblast cell lines from Cockayne syndrome (CS) patients, of either the CS-A or CS-B complementation groups, which are defective for pol II-mediated transcription-coupled DNA repair (TCR). It is not yet clear if the observed accumulation of ubiquitinated forms of the large subunit reflects a stimulation of ubiquitin-mediated degradation or if the ubiquitination serves a regulatory function independent of proteolysis. In either case, these results suggest that ubiquitination of the large subunit of pol II might be linked to its function in TCR. Experiments are underway to determine whether Rsp5 functions in the transcription-coupled repair pathway in yeast.
Rsp5 contains three “WW” motifs amino-terminal to the hect domain. WW domains consist of ≈38 amino acids and have been found in many proteins, including dystrophin and the 65-kDa yes-associated protein (YAP65) (22). It has been proposed that WW domains are protein-binding modules that recognize proline-rich elements containing a PXY motif. Interestingly, the CTD heptapeptide repeat consensus sequence is SPTSPSY, suggesting that the WW domains of Rsp5 might directly mediate the interaction with the heptapeptide repeats. Our experiments, however, have not ruled out the possibility that auxiliary factors might be involved in mediating the Rsp5/Rpb1 interaction; Bul1, for example, is a yeast protein that has been suggested to be a modulator of Rsp5 activity (23). Mouse Nedd4 and human RPF1 are hect domain proteins which, based on sequence similarity, might be functional homologs of Rsp5 (24, 25). Like Rsp5, Nedd4 and RPF1 contain WW domains within their amino-terminal regions. Experiments are underway to determine if these proteins target the largest subunit of mammalian pol II.
A genetic link between RSP5 and the basal transcription machinery was suggested previously by the fact that mutations in RSP5 were isolated in a search for suppressors of mutations in SPT3, which encodes a transcriptional activator that interacts with the TATA-binding component of TFIID (12). While the biochemical basis of this finding is not yet clear, one possibility is that spt3 mutations can be compensated for by a decrease in Rpb1 ubiquitination that would be conferred by an rsp5 mutant. It has also been shown that RSP5 and human RPF1 can potentiate hormone-dependent activation of transcription by the human progesterone receptor in yeast (25), which might also reflect a link between Rsp5 and the general transcriptional machinery.
Because RSP5 is an essential gene, we hypothesize that the lack of ubiquitination of one or more substrates of Rsp5 leads to cell inviability. We do not yet know if Rpb1 is the substrate of Rsp5 that is related to the essential function of Rsp5. Two other substrates of Rsp5 (Fur4 and Gap1) have been reported (11), and the experiment shown in Fig. 1 suggests that there may be additional substrates (p160, p60). Fur4 and Gap1 are both plasma membrane-associated permeases, although it has been suggested that targeting of these proteins is not related to the essential function of Rsp5 (11). The Schizosaccharomyces pombe homolog of Rsp5, Pub1, has been shown to target the Cdc25 protein phosphatase (26), and it appears that Nedd4 might target mammalian epithelial sodium channel (ENaC) subunits for ubiquitin-mediated degradation (27). Rsp5, Nedd4, and Pub1 all have a putative calcium-dependent phospholipid binding domain (C2 domain) near their amino terminus, which might be important in targeting membrane-associated substrates (28). Our preliminary structure/function analyses of Rsp5 indicate that this domain is not necessary for Rpb1 targeting.
All of the hect E3 proteins consist of a large amino-terminal domain and a conserved carboxyl-terminal hect domain. The region of Rsp5 involved in binding Rpb1 is within its amino-terminal domain, and the hect domain of Rsp5 contains all of the determinants necessary to accept ubiquitin from the E2 protein in the form of a thioester. These results are consistent with a two-domain model for hect E3 function in which the divergent amino-terminal domains direct substrate specificity, while the hect domain catalyzes ubiquitination of bound substrates. Further characterization of the enzyme/substrate interactions of the hect E3s will yield insight into the factors that control their substrate specificity as well as the processes and pathways that are affected by this class of proteins.
Acknowledgments
We thank Danny Reinberg and members of his lab, Fred Winston, and Steve Brill for supplying reagents and for helpful discussions, and Bill Lane and Harvard Microchemistry for peptide isolation and sequencing. We also thank Peter Howley for his support in the initial stages of this work, and Robert Krug, Mike Kaledjian, and Danny Reinberg for critical reading of the manuscript. This work was supported by a grant to J.M.H. from the American Cancer Society and from funds provided by Rutgers University.
ABBREVIATIONS
- CTD
carboxyl-terminal domain
- GST
glutathione S-transferase
- PVDF
polyvinylidene fluoride
- pol II
polymerase II
References
- 1.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 3.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M, Nuber U, Huibregtse J M. Nature (London) 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 7.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel B, Wünning I, Varshavsky A. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 10.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. Mol Micro. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 12.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 13.Hatfield P M, Callis J, Vierstra R D. J Biol Chem. 1990;265:15813–15817. [PubMed] [Google Scholar]
- 14.Girod P-A, Carpenter T P, van Nocker S, Sullivan M L, Vierstra R D. Plant J. 1993;3:545–552. doi: 10.1046/j.1365-313x.1993.03040545.x. [DOI] [PubMed] [Google Scholar]
- 15.Silver P A, Chiang A, Sadler I. Genes Dev. 1988;2:707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- 16.Allison L A, Wong K-C, Fitzpatrick V D, Moyle M, Ingles C J. Mol Cell Biol. 1988;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brill S J, Stillman B. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien T, Hardin S, Greenleaf A, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 19.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 20.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:133–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 21.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. Proc Natl Acad Sci USA. 1996;93:11, 586–11,590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 23.Yashiroda H, Oguchi T, Yasuda Y, Akio T-E, Kikuchi Y. Mol Cell Biol. 1996;16:3255–3263. doi: 10.1128/mcb.16.7.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Tomooka Y, Noda M. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 25.Imhof M O, McDonnell D P. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nefsky B, Beach D. EMBO J. 1996;15:1301–1312. [PMC free article] [PubMed] [Google Scholar]
- 27.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 28.Clark J D, Lin L L, Kriz R W, Ramesha C S, Sultzman L A, Lin A Y, Milona N, Knopf J L. Cell. 1991;65:1043–1051. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]