Abstract
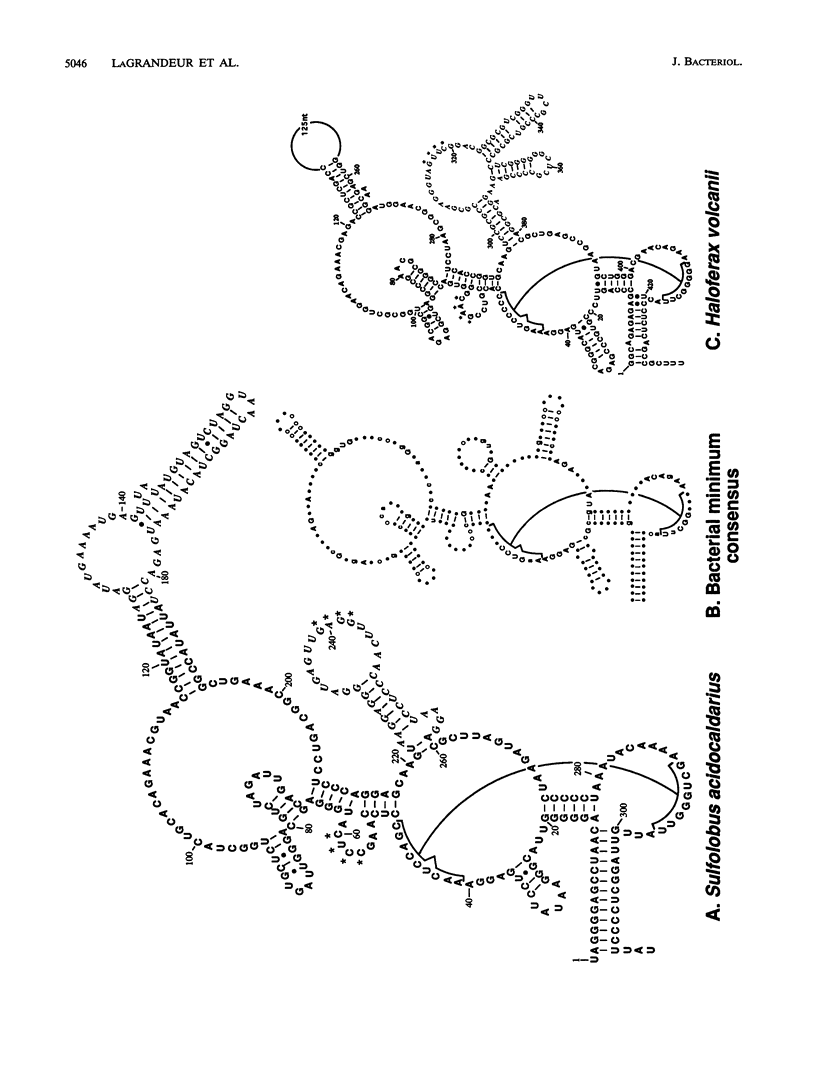
RNase P is the ribonucleoprotein enzyme that cleaves precursor sequences from the 5' ends of pre-tRNAs. In Bacteria, the RNA subunit is the catalytic moiety. Eucaryal and archaeal RNase P activities copurify with RNAs, which have not been shown to be catalytic. We report here the analysis of the RNase P RNA from the thermoacidophilic archaeon Sulfolobus acidocaldarius. The holoenzyme was highly purified, and extracted RNA was used to identify the RNase P RNA gene. The nucleotide sequence of the gene was determined, and a secondary structure is proposed. The RNA was not observed to be catalytic by itself, but it nevertheless is similar in sequence and structure to bacterial RNase P RNA. The marked similarity of the RNase P RNA from S. acidocaldarius and that from Haloferax volcanii, the other known archael RNase P RNA, supports the coherence of Archaea as a phylogenetic domain.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv Enzymol Relat Areas Mol Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- Baer M., Nilsen T. W., Costigan C., Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990 Jan 11;18(1):97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Haas E. S., James B. D., Hunt D. A., Liu J. S., Pace N. R. Phylogenetic analysis and evolution of RNase P RNA in proteobacteria. J Bacteriol. 1991 Jun;173(12):3855–3863. doi: 10.1128/jb.173.12.3855-3863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Darr S. C., Pace B., Pace N. R. Characterization of ribonuclease P from the archaebacterium Sulfolobus solfataricus. J Biol Chem. 1990 Aug 5;265(22):12927–12932. [PubMed] [Google Scholar]
- Doria M., Carrara G., Calandra P., Tocchini-Valentini G. P. An RNA molecule copurifies with RNase P activity from Xenopus laevis oocytes. Nucleic Acids Res. 1991 May 11;19(9):2315–2320. doi: 10.1093/nar/19.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Söll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Jul;5(7):1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Engelke D. R. Partial characterization of an RNA component that copurifies with Saccharomyces cerevisiae RNase P. Mol Cell Biol. 1989 Jun;9(6):2536–2543. doi: 10.1128/mcb.9.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwlandt D. T., Haas E. S., Daniels C. J. The RNA component of RNase P from the archaebacterium Haloferax volcanii. J Biol Chem. 1991 Mar 25;266(9):5689–5695. [PubMed] [Google Scholar]
- Pace N. R., Smith D. Ribonuclease P: function and variation. J Biol Chem. 1990 Mar 5;265(7):3587–3590. [PubMed] [Google Scholar]
- Reich C., Gardiner K. J., Olsen G. J., Pace B., Marsh T. L., Pace N. R. The RNA component of the Bacillus subtilis RNase P. Sequence, activity, and partial secondary structure. J Biol Chem. 1986 Jun 15;261(17):7888–7893. [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992 Jul 3;257(5066):74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S., Gamulin V., Burkard U., Söll D. The RNA component of RNase P in Schizosaccharomyces species. FEBS Lett. 1990 Oct 1;271(1-2):189–193. doi: 10.1016/0014-5793(90)80403-6. [DOI] [PubMed] [Google Scholar]