Abstract
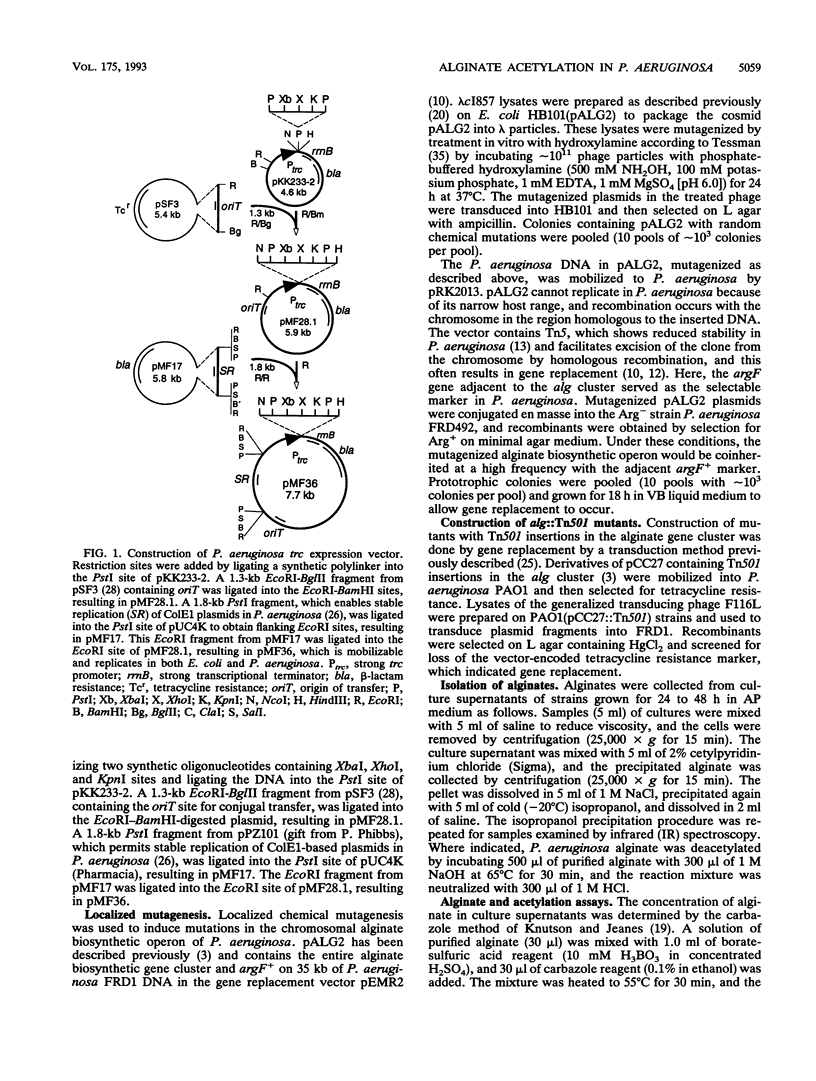
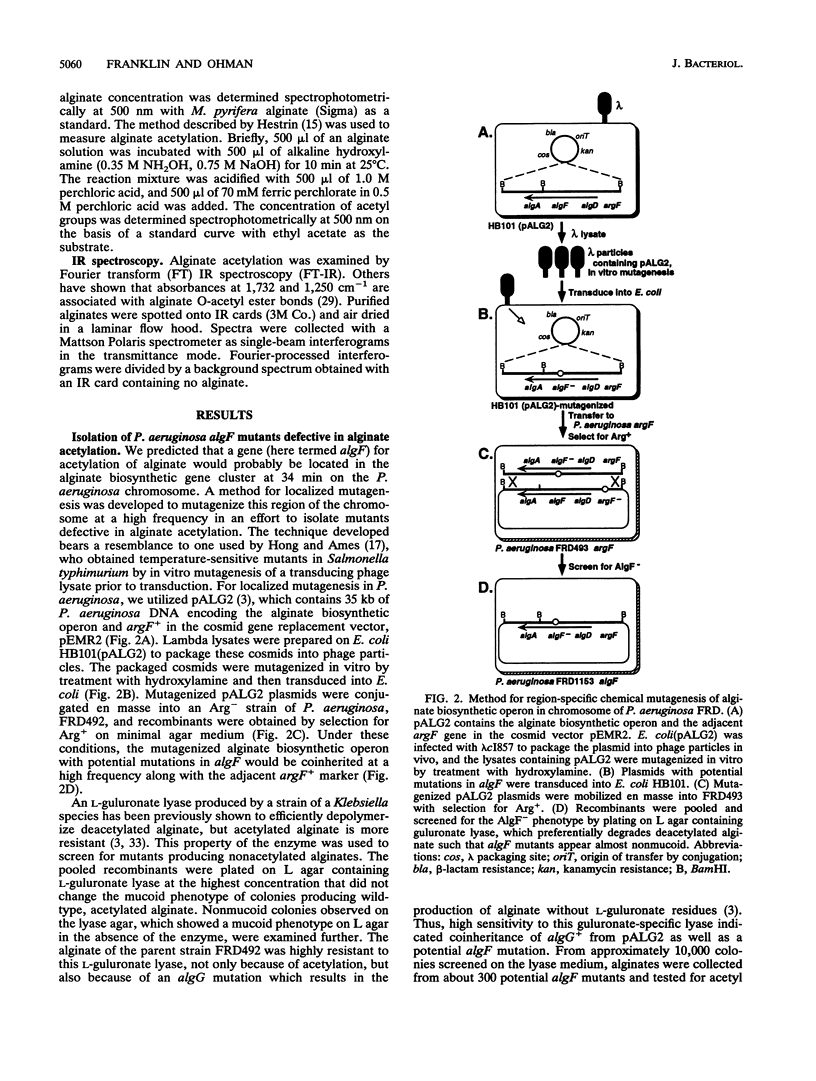
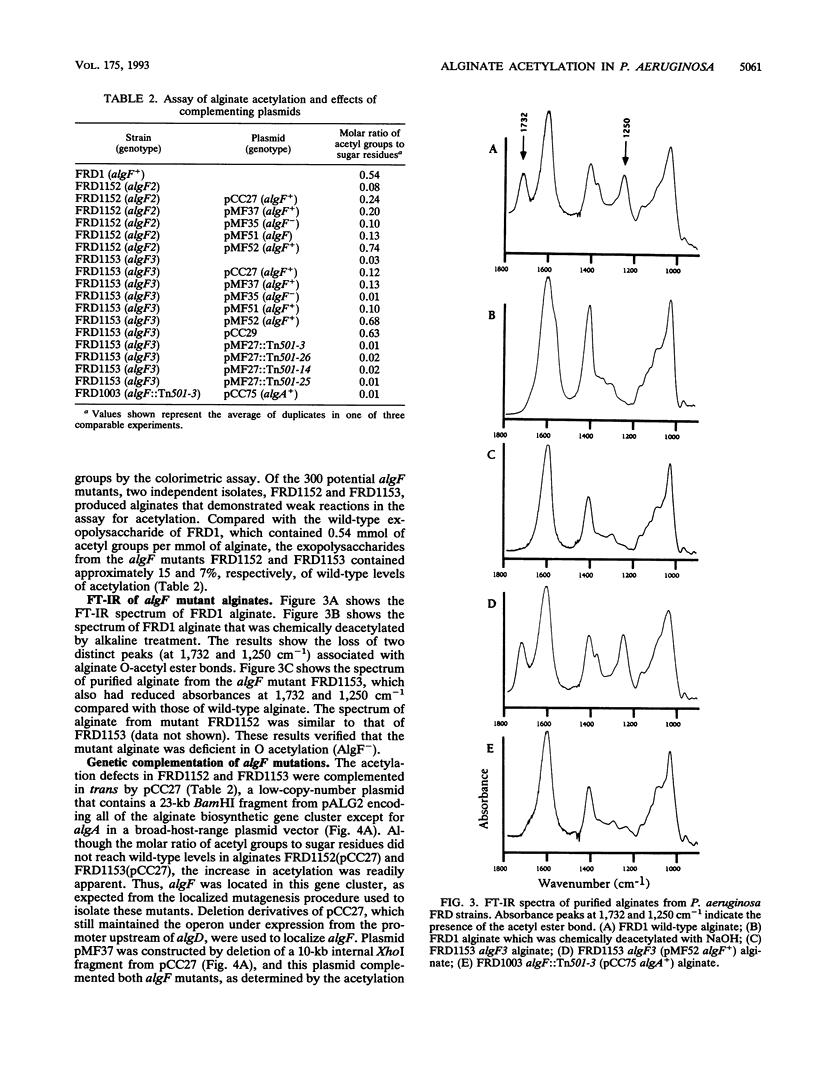
Mucoid strains of Pseudomonas aeruginosa produce a high-molecular-weight exopolysaccharide called alginate that is modified by the addition of O-acetyl groups. To better understand the acetylation process, a gene involved in alginate acetylation called algF was identified in this study. We hypothesized that a gene involved in alginate acetylation would be located within the alginate biosynthetic gene cluster at 34 min on the P. aeruginosa chromosome. To isolate algF mutants, a procedure for localized mutagenesis was developed to introduce random chemical mutations into the P. aeruginosa alginate biosynthetic operon on the chromosome. For this, a DNA fragment containing the alginate biosynthetic operon and adjacent argF gene in a gene replacement cosmid vector was utilized. The plasmid was packaged in vivo into lambda phage particles, mutagenized in vitro with hydroxylamine, transduced into Escherichia coli, and mobilized to an argF auxotroph of P. aeruginosa FRD. Arg+ recombinants coinherited the mutagenized alginate gene cluster and were screened for defects in alginate acetylation by testing for increased sensitivity to an alginate lyase produced by Klebsiella aerogenes. Alginates from recombinants which showed increased sensitivity to alginate lyase were tested for acetylation by a colorimetric assay and infrared spectroscopy. Two algF mutants that produced alginates reduced more than sixfold in acetyl groups were obtained. The acetylation defect was complemented in trans by a 3.8-kb XbaI-BamHI fragment from the alginate gene cluster when placed in the correct orientation under a trc promoter. By a merodiploid analysis, the algF gene was further mapped to a region directly upstream of algA by examining the polar effect of Tn501 insertions. By gene replacement, DNA with a Tn501 insertion directly upstream of algA was recombined with the chromosome of mucoid strain FRD1. The resulting strain, FRD1003, was nonmucoid because of the polar effect of the transposon on the downstream algA gene. By providing algA in trans under the tac promoter, FRD1003 produced nonacetylated alginate, indicating that the transposon was within or just upstream of algF. These results demonstrated that algF, a gene involved in alginate acetylation, is located directly upstream of algA.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore R. S., Mitchell M. Immunologic investigations of mucoid strains of Pseudomonas aeruginosa: comparison of susceptibility to opsonic antibody in mucoid and nonmucoid strains. J Infect Dis. 1980 Feb;141(2):238–247. doi: 10.1093/infdis/141.2.238. [DOI] [PubMed] [Google Scholar]
- Bolister N., Basker M., Hodges N. A., Marriott C. The diffusion of beta-lactam antibiotics through mixed gels of cystic fibrosis-derived mucin and Pseudomonas aeruginosa alginate. J Antimicrob Chemother. 1991 Mar;27(3):285–293. doi: 10.1093/jac/27.3.285. [DOI] [PubMed] [Google Scholar]
- Chitnis C. E., Ohman D. E. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990 Jun;172(6):2894–2900. doi: 10.1128/jb.172.6.2894-2900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis C. E., Ohman D. E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993 May;8(3):583–593. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- Chu L., May T. B., Chakrabarty A. M., Misra T. K. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene. 1991 Oct 30;107(1):1–10. doi: 10.1016/0378-1119(91)90290-r. [DOI] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. R., Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973 Nov;116(2):915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J Bacteriol. 1988 Jul;170(7):3228–3236. doi: 10.1128/jb.170.7.3228-3236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J Bacteriol. 1987 Apr;169(4):1593–1602. doi: 10.1128/jb.169.4.1593-1602.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Won J., Ohman D. E. Precise excision and instability of the transposon Tn5 in Pseudomonas aeruginosa. J Gen Microbiol. 1990 May;136(5):789–796. doi: 10.1099/00221287-136-5-789. [DOI] [PubMed] [Google Scholar]
- Govan J. R., Harris G. S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986 Oct;3(10):302–308. [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn J. M., Ohman D. E. Transcriptional and translational analyses of recA mutant alleles in Pseudomonas aeruginosa. J Bacteriol. 1988 Apr;170(4):1637–1650. doi: 10.1128/jb.170.4.1637-1650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson C. A., Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968 Sep;24(3):470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- Marcus H., Baker N. R. Quantitation of adherence of mucoid and nonmucoid Pseudomonas aeruginosa to hamster tracheal epithelium. Infect Immun. 1985 Mar;47(3):723–729. doi: 10.1128/iai.47.3.723-729.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Chakrabarty A. M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981 Jul;33(1):142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Chitnis C. E. Genetic regulation of alginate structure in Pseudomonas aeruginosa. Antibiot Chemother (1971) 1989;42:56–61. doi: 10.1159/000417604. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzmann S., Boring J. R. Antiphagocytic Effect of Slime from a Mucoid Strain of Pseudomonas aeruginosa. Infect Immun. 1971 Jun;3(6):762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj G., Fong Y. C., Iyer V. N. A portable DNA sequence carrying the cohesive site (cos) of bacteriophage lambda and the mob (mobilization) region of the broad-host-range plasmid RK2: a module for the construction of new cosmids. Gene. 1984 Dec;32(1-2):235–241. doi: 10.1016/0378-1119(84)90051-9. [DOI] [PubMed] [Google Scholar]
- Sherbrock-Cox V., Russell N. J., Gacesa P. The purification and chemical characterisation of the alginate present in extracellular material produced by mucoid strains of Pseudomonas aeruginosa. Carbohydr Res. 1984 Dec 15;135(1):147–154. doi: 10.1016/0008-6215(84)85012-0. [DOI] [PubMed] [Google Scholar]
- Shinabarger D., Berry A., May T. B., Rothmel R., Fialho A., Chakrabarty A. M. Purification and characterization of phosphomannose isomerase-guanosine diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991 Feb 5;266(4):2080–2088. [PubMed] [Google Scholar]
- Simpson J. A., Smith S. E., Dean R. T. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988 Jan;134(1):29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- Skjåk-Braek G., Grasdalen H., Larsen B. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr Res. 1986 Oct 15;154:239–250. doi: 10.1016/s0008-6215(00)90036-3. [DOI] [PubMed] [Google Scholar]
- Tessman I. Mutagenic treatment of double- and single-stranded DNA phages T4 ans S13 with hydroxylamine. Virology. 1968 Jun;35(2):330–333. doi: 10.1016/0042-6822(68)90275-4. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


