Abstract
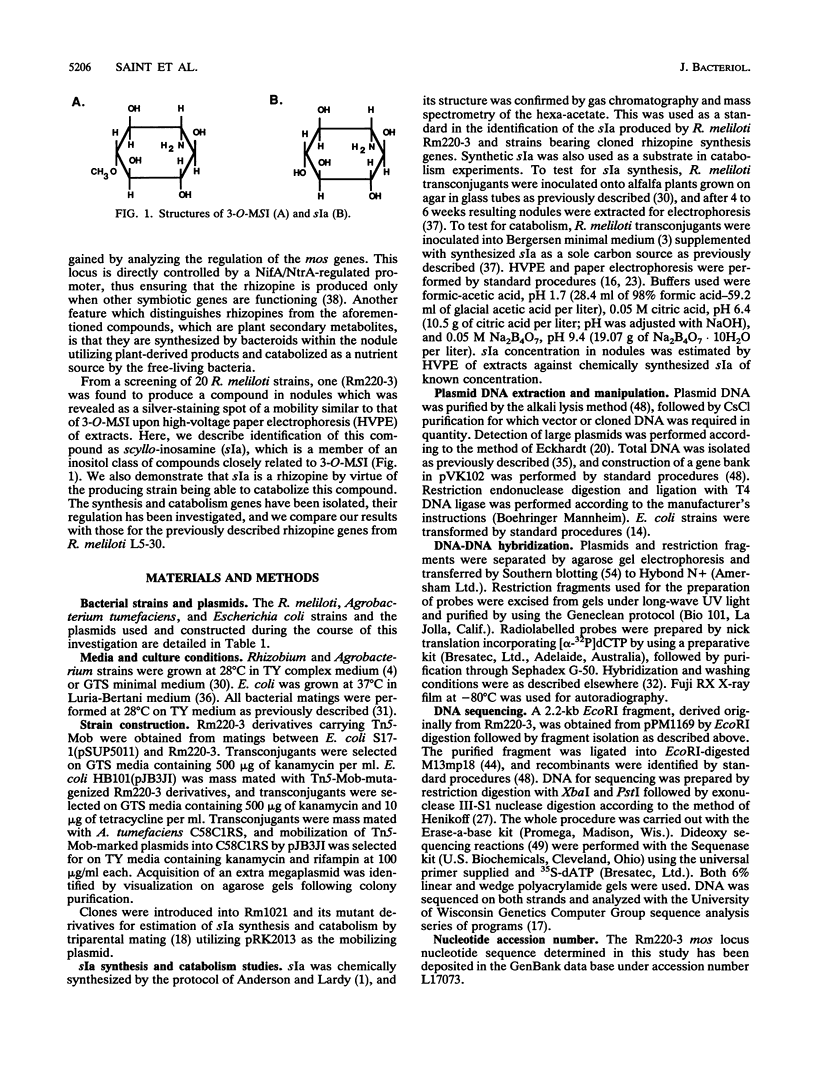
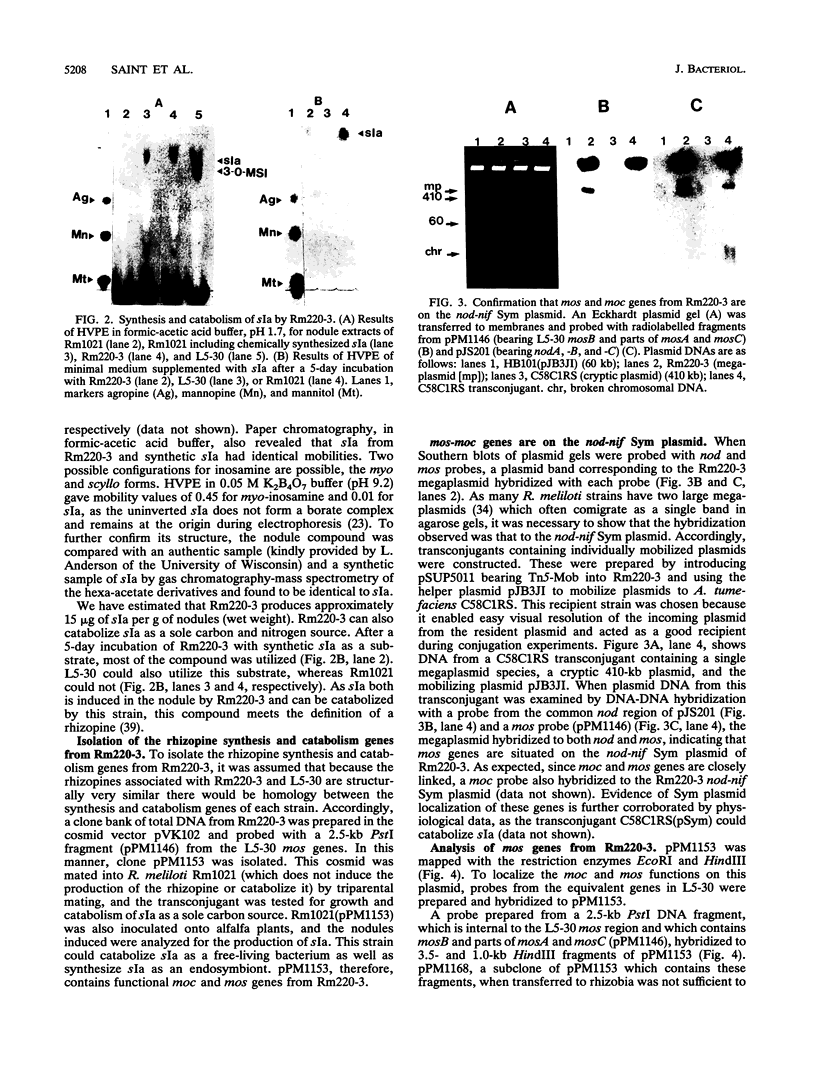
Rhizopines are selective growth substrates synthesized in nodules only by strains of rhizobia capable of their catabolism. We report the isolation and study of genes for the synthesis and catabolism of a new rhizopine, scyllo-inosamine (sIa), from alfalfa nodules induced by Rhizobium meliloti Rm220-3. This compound is similar in structure to the previously described rhizopine 3-O-methyl-scyllo-inosamine from R. meliloti L5-30 (P.J. Murphy, N. Heycke, Z. Banfalvi, M.E. Tate, F.J. de Bruijn, A. Kondorosi, J. Tempé, and J. Schell, Proc. Natl. Acad. Sci. USA 84:493-497, 1987). The synthesis (mos) and catabolism (moc) genes for the Rm220-3 rhizopine are closely linked and located on the nod-nif Sym plasmid. The mos genes are directly controlled by the NifA/NtrA regulatory system. A comparison of the sequence of the 5' regions of the two mos loci shows very extensive conservation of sequence as well as strong homology to the nifH coding region. Restriction mapping and hybridization to DNA from the four open reading frames (ORFs) of the L5-30 mos locus indicate the absence of mosA and presence of the other three ORFs (ORF1 and mosB and -C) in Rm220-3. We suggest that the L5-30 mosA gene product is involved in the conversion of scyllo-inosamine to 3-O-methyl-scyllo-inosamine. Restriction fragment length polymorphism analysis of the moc regions of both strains shows that they are very similar. Regulation studies indicate that the moc region is not controlled by the common regulatory gene nifA, ntrA, and ntrC. We discuss the striking similarities in gene structure, location, and regulation between these two rhizopine loci in relation to the rhizopine concept.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beringer J. E., Brewin N., Johnston A. W., Schulman H. M., Hopwood D. A. The Rhizobium--legume symbiosis. Proc R Soc Lond B Biol Sci. 1979 Apr 11;204(1155):219–233. doi: 10.1098/rspb.1979.0024. [DOI] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Better M., Lewis B., Corbin D., Ditta G., Helinski D. R. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983 Dec;35(2 Pt 1):479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- Boivin C., Barran L. R., Malpica C. A., Rosenberg C. Genetic analysis of a region of the Rhizobium meliloti pSym plasmid specifying catabolism of trigonelline, a secondary metabolite present in legumes. J Bacteriol. 1991 May;173(9):2809–2817. doi: 10.1128/jb.173.9.2809-2817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin C., Camut S., Malpica C. A., Truchet G., Rosenberg C. Rhizobium meliloti Genes Encoding Catabolism of Trigonelline Are Induced under Symbiotic Conditions. Plant Cell. 1990 Dec;2(12):1157–1170. doi: 10.1105/tpc.2.12.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Clare B. G., Kerr A., Jones D. A. Characteristics of the nopaline catabolic plasmid in Agrobacterium strains K84 and K1026 used for biological control of crown gall disease. Plasmid. 1990 Mar;23(2):126–137. doi: 10.1016/0147-619x(90)90031-7. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin D., Barran L., Ditta G. Organization and expression of Rhizobium meliloti nitrogen fixation genes. Proc Natl Acad Sci U S A. 1983 May;80(10):3005–3009. doi: 10.1073/pnas.80.10.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. N., Broughton W. J. Competition for nodulation of legumes. Annu Rev Microbiol. 1986;40:131–157. doi: 10.1146/annurev.mi.40.100186.001023. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Hartwig U. A., Joseph C. M., Phillips D. A. Flavonoids Released Naturally from Alfalfa Seeds Enhance Growth Rate of Rhizobium meliloti. Plant Physiol. 1991 Mar;95(3):797–803. doi: 10.1104/pp.95.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Heusterspreute M., Ha Thi V., Emery S., Tournis-Gamble S., Kennedy N., Davison J. Vectors with restriction site banks. IV. pJRD184, a 3793-bp plasmid vector with 49 unique restriction sites. Gene. 1985;39(2-3):299–304. doi: 10.1016/0378-1119(85)90327-0. [DOI] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kowalski M. Transduction in Rhizobium meliloti. Acta Microbiol Pol. 1967;16(1):7–11. [PubMed] [Google Scholar]
- Long S. R. Rhizobium genetics. Annu Rev Genet. 1989;23:483–506. doi: 10.1146/annurev.ge.23.120189.002411. [DOI] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P. J., Heycke N., Banfalvi Z., Tate M. E., de Bruijn F., Kondorosi A., Tempé J., Schell J. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc Natl Acad Sci U S A. 1987 Jan;84(2):493–497. doi: 10.1073/pnas.84.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P. J., Heycke N., Trenz S. P., Ratet P., de Bruijn F. J., Schell J. Synthesis of an opine-like compound, a rhizopine, in alfalfa nodules is symbiotically regulated. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9133–9137. doi: 10.1073/pnas.85.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
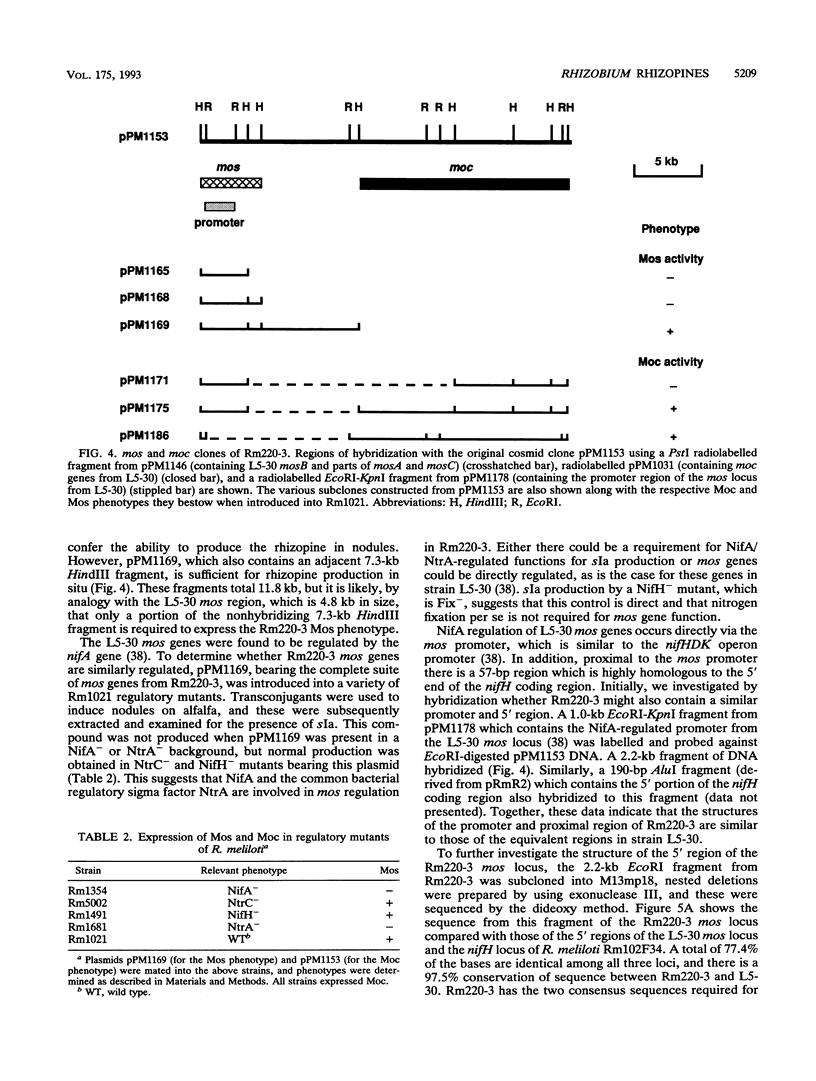
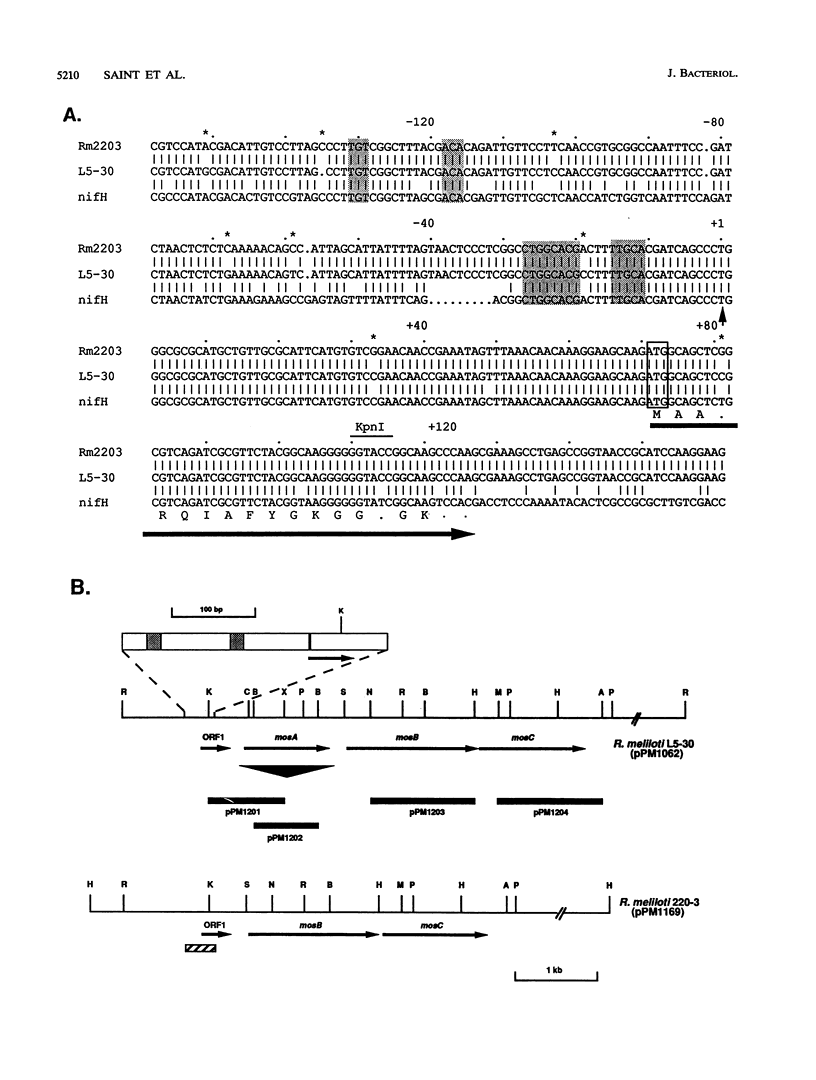
- Murphy P. J., Trenz S. P., Grzemski W., De Bruijn F. J., Schell J. The Rhizobium meliloti rhizopine mos locus is a mosaic structure facilitating its symbiotic regulation. J Bacteriol. 1993 Aug;175(16):5193–5204. doi: 10.1128/jb.175.16.5193-5204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal C. S., Dion P., Chilton W. S. Mannopine and mannopinic acid as substrates for Arthrobacter sp. strain MBA209 and Pseudomonas putida NA513. J Bacteriol. 1991 May;173(9):2833–2841. doi: 10.1128/jb.173.9.2833-2841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Albright L. M., Ausubel F. M. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol. 1987 Jun;169(6):2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Sundaresan V., Ausubel F. M. Directed transposon Tn5 mutagenesis and complementation analysis of Rhizobium meliloti symbiotic nitrogen fixation genes. Cell. 1982 Jun;29(2):551–559. doi: 10.1016/0092-8674(82)90171-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., John M., Kondorosi E., Kondorosi A., Wieneke U., Schröder G., Schröder J., Schell J. Mapping of the protein-coding regions of Rhizobium meliloti common nodulation genes. EMBO J. 1984 Aug;3(8):1705–1711. doi: 10.1002/j.1460-2075.1984.tb02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. B., Wilson R., Shaw G. J., Petit A., Tempe J. Biosynthesis and degradation of nodule-specific Rhizobium loti compounds in Lotus nodules. J Bacteriol. 1987 Jan;169(1):278–282. doi: 10.1128/jb.169.1.278-282.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196(3):413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Szeto W. W., Nixon B. T., Ronson C. W., Ausubel F. M. Identification and characterization of the Rhizobium meliloti ntrC gene: R. meliloti has separate regulatory pathways for activation of nitrogen fixation genes in free-living and symbiotic cells. J Bacteriol. 1987 Apr;169(4):1423–1432. doi: 10.1128/jb.169.4.1423-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto W. W., Zimmerman J. L., Sundaresan V., Ausubel F. M. A Rhizobium meliloti symbiotic regulatory gene. Cell. 1984 Apr;36(4):1035–1043. doi: 10.1016/0092-8674(84)90053-9. [DOI] [PubMed] [Google Scholar]
- Thöny B., Hennecke H. The -24/-12 promoter comes of age. FEMS Microbiol Rev. 1989 Dec;5(4):341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Tremblay G., Gagliardo R., Chilton W. S., Dion P. Diversity among Opine-Utilizing Bacteria: Identification of Coryneform Isolates. Appl Environ Microbiol. 1987 Jul;53(7):1519–1524. doi: 10.1128/aem.53.7.1519-1524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]