Abstract
The N-terminal 70 residue “J-domain” of the Escherichia coli DnaJ molecular chaperone is the defining and highly conserved feature of a large protein family. Based upon limited, yet significant, amino acid sequence homology to the J-domain, the DNA encoding the T/t common exon of the simian virus 40 (SV40), JC, or BK polyoma virus T antigen oncoproteins was used to construct J-domain replacement chimeras of the E. coli DnaJ chaperone. The virally encoded J-domains successfully substituted for the bacterial counterpart in vivo as shown by (i) complementation for viability at low and high temperature of a hypersensitive bacterial reporter strain, and (ii) the restoration of bacteriophage λ plaque forming ability in the same strain. The amino acid change, H42Q, in the SV40 T/t and the JC virus T/t exon, which is positionally equivalent to the canonical dnaJ259 H33Q mutation within the E. coli J-domain, entirely abolished complementing activity. These results strongly suggest that the heretofore functionally undefined viral T/t common exon represents a bona fide J-domain that preserves critical features of the characteristic domain fold essential for J-domain interaction with the ATPase domain of the Hsp70 family. This finding has implications for the regulation of DNA tumor virus T antigens by molecular chaperones.
Keywords: JC virus, BK virus, Hsp70, DnaK, Hsp40
The DnaJ family of molecular chaperones is a key regulator of protein folding, assembly, and transport in both eukaryotes and prokaryotes (1–4). A highly conserved, 70-residue “J-domain,” the defining feature of this large and evolutionarily diverse family, is implicated in the modulation of Hsp70 family chaperone activity. Considerable evidence exists to suggest that the J-domain functions, in part, by direct interaction with Hsp70’s ATPase domain to stimulate ATP hydrolysis, and thereby concomitantly modulate Hsp70 conformational states and substrate binding/release during the Hsp70 chaperone cycle (5–12).
Simian virus 40 (SV40) has been extensively studied as a model DNA tumor virus (13). Human polyoma viruses, JC virus (JCV) and BK virus (BKV), are highly homologous to SV40 (14). JCV is widespread and asymptomatic in the human population, though in rare cases, latent virus can reactivate under immunosuppressed conditions to cause a fatal demyelinating brain disease, termed progressive multifocal leukoencephalopathy. BKV is equally widespread in the population, though its pathology is less well defined (15).
Alternative splicing of the polyoma virus family early message generates large T antigen (T), small t antigen (t), and in murine and hamster polyoma, middle-T antigen (mt) (13). The T/t common exon, coding for residues residues 1–82, is retained in all early spliced gene products and forms the extreme N terminus in nearly all the viral early proteins. These early proteins play numerous roles in the viral life cycle. SV40 T is a complex oncoprotein shown to have domains for DNA binding and ATPase-helicase, DNA polymerase α association, binding of p53 and members of the retinoblastoma (pRB) anti-oncogene family, and binding of transcription preinitiation complex factors (13, 16–21). T is also regulated by specific phosphorylation and self-oligomerization (22, 23). SV40 t can bind protein phosphatase 2A and can help promote cellular transformation by modulation of mitogen-activated protein kinase signaling pathways (24), while polyoma mt can associate with members of the src family of tyrosine kinases and control aspects of cell proliferation (25). Recent work has implicated the N-terminal T/t common domain of SV40 T in the control of cell cycle and growth regulatory interactions imposed by p53, the pRB-related proteins p107 and p130, and the transcriptional coactivator proteins p300 and CBP (21, 26–30). Further studies reveal a role for the N-terminal domain in transcriptional activation and repression (31, 32), and in immortalization, transformation, DNA replication, protein stability, and viral morphogenesis (33–39). Despite intensive research, a definitive understanding of the function of the T/t common domain in each of these viral early proteins has remained elusive.
We have previously noted a limited, yet significant, sequence similarity among the polyoma viral family T/t common domain and the J-domains from diverse organisms (ref. 40 and Fig. 1). Here, we present multiple lines of evidence showing that the T/t common domain of SV40, JCV, and BKV can functionally substitute for the J-domain of the E. coli DnaJ molecular chaperone in vivo.
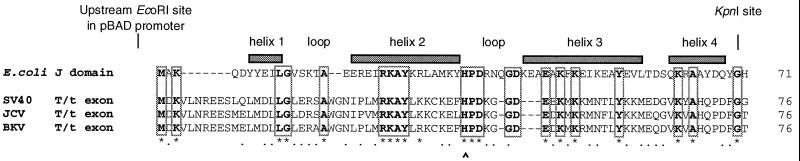
Figure 1.
Alignment of the E. coli DnaJ molecular chaperone J-domain and the viral T antigen N-terminal domains used in this study. Alignment to residue 76 is shown, as no homology was noted up to the exon splice junction site at residue 82. Helices span segments 6–10, 18–32, 41–57, and 61–68 and an antiparallel coiled coil composed of helices 2–3 and anchored by helix 1 form the essential structural fold of the E. coli J-domain (41–43). A circumflex (^) shows the location of the mutation H33Q (ref. 11; E. coli dnaJ259) and the positionally equivalent mutation, H42Q, in the viral sequences. Identical residues have been highlighted in bold and boxed. The restriction sites used for the construction of the chimeras are shown.
MATERIALS AND METHODS
Bacterial Strains.
CU247 (44), LMG190 (45) containing Δara714, and MC4100 (46) have been described. WKG190 = MC4100, araD139 Δara714 ΔcbpA::kan dnaJ::Tn10-42 and was made by successive P1 transductions using MC4100, CU247, and LMG190. WKG191, WKG192, WKG193, WKG194, WKG195, and WKG196 are MC4100 araD139 Δara714 derivatives constructed by P1- or T4 gt7-transduction and carrying, respectively, ΔdnaK52::cam sidB1 (47), dnaK103 thr::Tn10 (48), grpE280 pheA::Tn10 (49), dnaK756 thr::Tn10 (48), dnaK103 thr::Tn10 ΔgrpE::Ωcam (48), ΔdnaK14 dnaJ14::kan (50).
DnaJ/DnaJ12 Expression Plasmid Construction.
A 1428-bp genomic E. coli DdeI–SmaI dnaJ+ fragment was cloned with the N-terminal adaptors (5′-GATCCATGGC-3′, 5′-TTAGCCATG-3′) into BamHI–EcoRV-cut pWKS30 (51) to give pWKG89a. The NcoI–XhoI fragment was then cut and cloned into a pBAD22 (45) derivative (pBAD22ΔHB2, HindIII to BglII) to yield pWKG90. The dnaJ12 expression plasmid was made by HindIII digestion of pWKG89a followed by religation of the vector fragment with the 883 bp dnaJ HindIII fragment containing the original dnaJ12 c-t transition (ref. 11; Q109stop) mutation. The dnaJ12 mutation itself introduced a HincII site so that HincII digestion at this site and the downstream polylinker site followed by religation of the vector introduced a TGA stop following the R108 codon and elimination of all downstream dnaJ coding sequence. The 329-bp dnaJ12 fragment was cut with NcoI–XhoI and cloned into NcoI–SalI cut pBAD22ΔHB2 to yield pWKG100.
PCR Construction of J-Domain Chimeras.
Primers A (5′-GGGAATTCACCATGGATAAAGTTTTAAAC-3′), B (5′-GGGATCCAGATCTTACGGTACCCTCAGTTGCATCCCAG-3′), C (5′-CCGGTACCAAAGTCAGGTTGATGAGC-3′), D (5′-GGGAATTCACCATGGACAAAGTGCTGAA-3′), E (5′-CCGGTACCAAAATCAGGCTGATGAGC-3′), and F (5′-GGGAATTCACCATGGATAAAGTTCTTAA-3′) were used as pair sets to amplify: SV40 1–82 (A plus B), SV40 1–76 (A plus C), JCV 1–76 (D plus E), BKV 1–76 (F plus E) J-domains using pUC19-SV40, JCV pMad1-TC, and pBKV-9 viral DNA templates (14).
Oligonucleotide-Directed Mutagenesis.
DnaJ/DnaJ12 expression phagemids were mobilized with VCSM13 (Stratagene) in strain CJ236 and mutagenesis was performed using the method of Kunkel (52). H71T was introduced into both pWKG90 and pWKG100 using the oligonucleotide 5′-CTCAAACGCAGCGGTACCATACTGATC-3′. All downstream residues from and including codon A72 were as in wild-type DnaJ. All KpnI–chimera junction sequences contained the H71T mutation preceded by viral coding sequence. Point mutants within the J-domain were prepared using the primers: dnaJ259-H33Q (5′-GTTACGGTCCGGTTGGTATTTCATG-3′), SV40–H42Q (5′-TCCTTTATCAGGTTGAAACTCCTTGCA-3′), JCV–H42Q (5′-ACCTTTATCAGGTTGGAGTTCTTTG-3′). All constructs were verified by dideoxy sequencing using Sequenase version 2.0 and appropriate primers.
Protein Stability Assay.
Fresh cultures were diluted 1:100 and grown to mid-log phase in Luria–Bertani broth containing l-arabinose (6.6 mM) and 50 μg/ml ampicillin at 30°C. At t = 0, protein synthesis was stopped with 30 μg/ml chloramphenicol. Aliquots were removed at intervals, whole-cell extracts prepared, and protein stability estimated from immunoblot analysis by enhanced chemiluminescence detection (Amersham) using rabbit polyclonal anti-DnaJ antibody and goat anti-rabbit IgG/horseradish peroxidase-conjugated secondary antibody (Bio-Rad).
Quantitative Protein Analysis.
Whole-cell extracts were normalized to A600 optical density and appropriate volume-corrected aliquots loaded on 12.5% SDS/PAGE gels, stained by Coomassie brilliant blue and relative proteins levels quantified by Cybertech digitized video image analysis and wincam 2.1 software. Purified DnaJ was used to establish linearity of the assay.
RESULTS
Establishment of an Assay for J-Domain Function.
An E. coli strain that lacks both dnaJ and its closely related homologue, cbpA, is hypersensitive for growth above 37°C and below 16°C and cannot support the growth of bacteriophage λ at any temperature (44). We have reconstructed this dnaJ cbpA double null strain in a genetic background that lacks the arabinose operon, but carries the araD139 allele to permit the use of l-arabinose as a nonmetabolized inducer molecule. Since this strain, WKG190, lacks all detectable DnaJ chaperone activity, complementation for growth at high or low temperature, or for permissivity to growth of bacteriophage λ, becomes dependent upon the conditional expression of DnaJ under the control of the arabinose-inducible pBAD promoter (45).
Two expression vector sets were constructed in parallel: one (1–376) contained the full length dnaJwt, while the other (1–108) used dnaJ12, a truncated version coding for only the J- and glycine-phenylalanine rich (G/F)-domain, yet still retaining many activities attributed to the full-length DnaJ despite loss of over 70% of the molecule (11, 12, 53, 54). The SV40, JCV, and BKV, T/t exons (1–82 or 1–76) were amplified by PCR and used to construct the J-domain replacement chimeras (Fig. 2). J-domain functional complementation in this assay system would predict that the viral J-domain chimeras should (i) complement for DnaJ activity by restoration of colony formation in the hypersensitive WKG190 reporter strain at both low and high temperature; (ii) support the growth of bacteriophage λ in WGK190, as λ specifically requires DnaJ activity at an early step of its DNA replication program (55, 56); and (iii) function in both the full-length 1–376 and minimal 1–108 DnaJ expression vector systems. We also reasoned that a single point mutation within the J-domain, positionally equivalent to the canonical dnaJ259 H33Q inactivating mutation (11), should also abolish activity of the chimeric DnaJ proteins, if indeed the viral T/t common exon were functioning mechanistically as a J-domain.
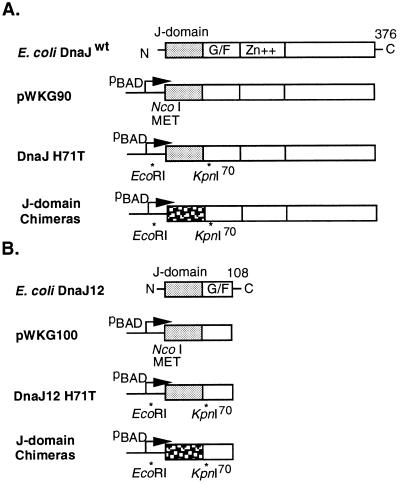
Figure 2.
Construction of J-domain PCR replacement chimeras. (A) Plasmids derived from the dnaJ wild-type (dnaJwt) clone. Four domains comprising the N-terminal J-domain, glycine-phenylalanine rich region (G/F), Zn+ finger domain, and a less conserved C-terminal domain are shown (11). (B) Plasmids derived from a dnaJ12 clone that lacks all coding sequence beyond residue 108. A phenotypically neutral mutation was introduced (H71T) that generated a unique KpnI site at the end of the J-domain. Viral T/t exons were PCR amplified to introduce the appropriate EcoRI and KpnI sites. The J-domain is shown stippled and proximal to the pBAD promoter used for conditional expression.
J-Domain Chimeras Complement in Vivo. A colony forming assay (Table 1 and Fig.
Table 1.
In vivo complementation for DnaJ chaperone activity by dnaJ plasmid chimeras carrying mammalian polyoma virus T antigen T/t common exons in place of the E. coli DnaJ chaperone J-domain
| Plasmid | dnaJ backbone | J-domain |
l-Arabinose inducer
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None
|
66 μM
|
660 μM
|
6.6 mM
|
33 mM
|
|||||||||||||
| 14.5 | 39.5 | 43 | 14.5 | 39.5 | 43 | 14.5 | 39.5 | 43 | 14.5 | 39.5 | 43 | 14.5 | 39.5 | 43 | |||
| pBAD22A vector | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| pWKG90 | J (71–376) | E. coli J(1-70) | - | - | - | - | 0.14 | - | 0.61 | 0.92 | 0.06 | 0.80 | 1.08 | 0.92 | 0.91 | 1.04 | 0.93 |
| pWKG92 | J (71–376) | SV40 (1-82) T/t | - | - | - | - | - | - | - | - | - | 0.83 | 1.00 | - | 1.17 | 0.03 | 0.03 |
| pWKG93 | J (71–376) | SV40 (1-76) | - | - | - | - | - | - | - | - | - | 0.54 | 0.87 | 0.03 | 0.83 | 0.07 | - |
| pWKG95 | J (71–376) | JCV (1-76) | - | - | - | - | - | - | - | - | - | 0.58 | 0.95 | 0.43 | 0.84 | 0.45 | 0.20 |
| pWKG97 | J (71–376) | BKV (1-76) | - | - | - | - | - | - | - | - | - | 0.11 | 0.94 | - | 0.90 | 0.36 | - |
| pWKG91 | J (71–376) | E. coli (1-70)H33Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| pWKG94 | J (71–376) | SV40 (1-76)H42Q | - | - | - | - | - | - | - | - | - | - | - | - | -* | -* | -* |
| pWKG96 | J (71–376) | JCV (1-76)H42Q | - | - | - | - | - | - | - | - | - | - | - | - | -* | -* | -* |
| pWKG100 | J (71–108) | E. coli J (1-70) | - | - | - | - | - | - | - | - | - | 0.65 | 0.66 | - | 0.84 | 0.85 | - |
| pWKG102 | J (71–108) | SV40 (1-82)T/t | - | - | - | - | - | - | - | - | - | 0.03 | - | - | 0.75 | 0.97 | - |
| pWKG103 | J (71–108) | SV40 (1-76) | - | - | - | - | - | - | - | - | - | 0.34 | - | - | 0.67 | 0.89 | - |
| pWKG105 | J (71–108) | JCV (1-76) | - | - | - | - | - | - | - | - | - | - | - | - | 0.57 | 1.03 | - |
| pWKG107 | J (71–108) | BKV (1-76) | - | - | - | - | - | - | - | - | - | - | - | - | 0.008 | 0.07 | - |
| pWKG101 | J (71–108) | E. coli (1-70)H33Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| pWKG104 | J (71–108) | SV40 (1-76)H42Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| pWKG106 | J (71–108) | JCV (1-76)H42Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
Colony formation at both low and high temperatures was measured using the host strain WKG190. WKG190 containing one of the indicated plasmids was grown at 30°C in Luria–Bertani (LB) broth containing 50 μg/ml ampicillin. Serial dilutions were spot tested on LB-ampicillin plates containing none, or the indicated amount of l-arabinose inducer and grown at the indicated temperature (14.5°C, 39.5°C, or 43°C). Cells were counted and efficiencies of plating were normalized to the viable cell titer of the same dilutions determined at 30°C. Plasmids carrying SV40–H42Q or JCV–H42Q mutations showed reduced viability at 30°C at 33 mM arabinose. The (-) sign indicates no detectable complementation [efficiency of plating (eop) < 10−5]. The values reported represent the mean of at least three independent determinations.
For certain plasmids, the addition of arabinose at high concentration resulted in a reduction of cell viability, even at 30°C.
3) strikingly revealed that all viral J-domain chimeras could complement for bacterial growth at 14.5°C and 39.5°C. The eop between 0.01 and 1.0 were observed provided that sufficient arabinose inducer was present. Control experiments showed that the pBAD22A vector alone was unable to complement under all conditions tested (eop < 10−5). On the contrary, the two positive controls, pWKG90 (dnaJwt) or pWKG100 (dnaJ12) could rescue bacterial growth at 14.5°C and 39.5°C with an eop of 0.05–1.0 even when 10-fold, or less, arabinose inducer was present than for the viral J-domain chimeras. Under no circumstances was pWKG100 or any of its dervatives observed to complement at 43°C.
Control experiments showed that the E. coli J-domain 1–70 alone, or the SV40 1–82 T/t exon alone were unable to complement under all conditions tested (data not shown). These results are in agreement with the previously published observation that the J-domain and the adjacent G/F domain represent the minimal truncation that permits retention of detectable DnaJ activity in E. coli (11, 12, 53, 54).
An Analogous Point Mutation Abolishes Complementing Activity.
An analogous point mutation that alters the highly conserved HPD residue cluster was introduced in the SV40 and JCV J-domain chimeras by site-directed mutagenesis. The failure of plasmids carrying the SV40–H42Q or JCV–H42Q J-domain chimeras to complement in this assay (eop <10−5) parallels the failure of plasmids carrying the canonical dnaJ259 H33Q mutation to complement under all conditions tested. This result argues forcefully in favor of the notion that the intrinsic mechanism of J-domain function has been retained in the viral T/t common domain and can be inactivated by a mutation positionally equivalent to the dnaJ259 H33Q (11). It is noteworthy that in other systems, dnaJ259-equivalent lesions YDJ1(H34Q) and MDJ1(H89Q) also display mutant phenotypes in Saccharomyces cerevisiae (10, 57) as are H to Q mutations in two additional proteins that contain J-domains only (unpublished data). The H residue lies in an exposed loop region of the J-domain and mutation there would not be expected to grossly alter the structure of the J-domain, as the domain fold is stabilized by many hydrophobic interactions on the inner faces of helices 1–3. Though it cannot be strictly eliminated, it is unlikely that the H to Q mutations alter the J-domain structure, but rather, inactivate a crucial solvent exposed residue necessary for interaction with the Hsp70 cognate partner ATPase domain.
Restoration of Bacteriophage λ Plaque Formation by J-Domain Chimeras.
Table 2 shows the results of a bacteriophage λ plaque-forming assay on strain WKG190 harboring the indicated dnaJ chimeric plasmids, or vector alone. The control λ dnaJ+ transducing bacteriophage was expected to form plaques with or without arabinose, whereas λ vir was expected to grow only on plates where both the arabinose inducer and a complementing plasmid providing DnaJ function were present. Titrations of arabinose revealed that concentrations that were not sufficient to support complementation for colony formation at high or low temperature (Table 1) were nevertheless sufficient to support growth of bacteriophage λ in this assay. The results clearly showed that the viral J-domain chimeras could complement for the DnaJ chaperone and restore the ability of λ vir to form plaques on strain WKG190. Plasmids containing the dnaJ259 H33Q, or the SV40–H42Q/JCV–H42Q mutations were unable to complement for bacteriophage growth under the conditions of this assay, in agreement with the results shown in Table 1.
Table 2.
Complementation assay for bacteriophage λ plaque formation at 30°C on strain WKG190
| plasmid | J-domain | pfu: no arabinose
|
pfu: 66 μmol l-arabinose
|
||
|---|---|---|---|---|---|
| λ dnaJ+ | λ vir | λ dnaJ+ | λ vir | ||
| pBAD22A vector | 0.28 | - | 0.17 | - | |
| pWKG90 | E. coli J(1-70) | 0.49 | 0.07 | 0.09 | 0.29 |
| pWKG92 | SV40 (1-82)T/t | 0.15 | - | 0.06 | 0.10 |
| pWKG93 | SV40 (1-76) | 0.27 | - | 0.09 | 0.35 |
| pWKG95 | JCV (1-76) | 0.29 | - | 0.20 | 0.11 |
| pWKG97 | BKV (1-76) | 0.26 | - | 0.12 | 0.14 |
| pWKG91 | E. coli J(1-70)H33Q | 0.14 | - | 0.003 | - |
| pWKG94 | SV40 (1-76)H42Q | 0.26 | - | 0.05 | - |
| pWKG96 | JCV (1-76)H42Q | 0.61 | - | 0.22 | - |
| pWKG100 | E. coli J(1-70) | 0.38 | - | 0.30 | 0.24 |
| pWKG102 | SV40 (1-82)T/t | 0.31 | - | 0.71 | 0.10 |
| pWKG103 | SV40 (1-76) | 0.27 | - | 0.27 | 0.11 |
| pWKG105 | JCV (1-76) | 0.23 | - | 0.21 | 0.15 |
| pWKG107 | BKV (1-76) | 0.20 | - | 0.16 | 0.02 |
| pWKG101 | E. coli J (1-70)H33Q | 0.11 | - | 0.14 | - |
| pWKG104 | SV40 (1-76)H42Q | 0.23 | - | 0.39 | - |
| pWKG106 | JCV (1-76)H42Q | 0.35 | - | 0.36 | - |
The control λ dnaJ+ transducing bacteriophage (λimm21 dnaJ+), or tester phage λ vir, were serially diluted and spot tested on bacterial lawns of WKG190 that carried the indicated chimeric dnaJ expression plasmids. The Luria–Bertani plates and soft agar overlays contained either none, or 66 μM l-arabinose as inducer, and 50 μg/ml ampicillin. For normalization, bacteriophage were also titered simultaneously on LMG190, an isogenic dnaJ+cbpA+ parent strain of WKG190. Normalized plaque forming units (pfu) are reported as the mean of at least three independent determinations. In certain cases, strains harboring plasmids with E. coli dnaJ259 (H33Q) or the viral equivalent mutations exhibit a partially dominant negative phenotype to dnaJ+, as shown by a reduction in its pfu efficiency, and/or plaque morphology. The (-) sign indicates no detectable complementation (eop < 10−5).
Functional complementation observed by expression of DnaJ and J-domain chimeras in strain WKG190 could possibly be the result of activation of an alternative mechanism that bypassed the requirement for DnaK (Hsp70) and the DnaK-specific nucleotide-release factor, GrpE. As a control, we constructed five strains, WKG191–WKG195, that contained missense or deletion alleles of dnaK and grpE, and a sixth strain, WKG196, deleted for both dnaK and dnaJ. Each strain was transformed with plasmids of the pWKG90 series and tested for rescue of the characteristic phenotypic defects associated with each allele. It turned out that neither DnaJ nor any of the chimeras could complement for the temperature-sensitive bacterial growth defects or the block to the productive growth of bacteriophage λ in these strains (data not shown). These results strongly argue that the DnaJ chimeras do not suppress bacterial and bacteriophage growth through a nonspecific mechanism; rather, the viral J-domain chimeras exert their effects in concert with the chaperone machine proteins DnaK and GrpE in the experiments reported in this work.
Analysis of Protein Levels and Protein Stability.
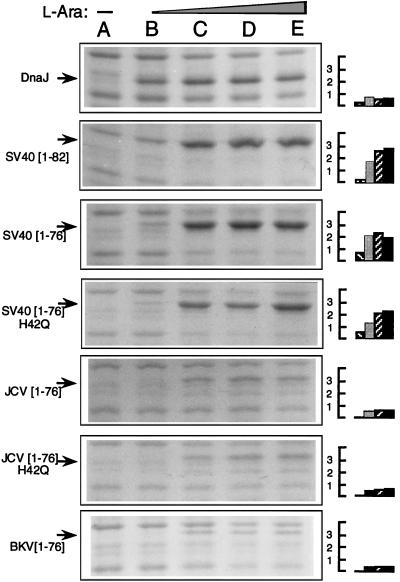
Variation in protein expression, stability, or amino acid differences in the J-domains could account for the need for more, or less, arabinose inducer in these assays. Quantitative protein analysis for the pWKG90 series of expression plasmids revealed minor variations in arabinose dose-response (Fig. 4). When comparing protein levels under conditions that yielded an eop of ≈1 in the temperature-sensitive complementation assay (660 μM arabinose for pWGK90; 6.6 mM for pWKG92, pWKG93, pWKG95, and pWKG97) we observed that the SV40 chimera protein levels were 2.5- to 3.7-fold higher than DnaJ, the JCV chimera levels were equivalent to DnaJ within the limits of experimental error, and the levels of BKV chimera were 1.5- to 2-fold lower than DnaJ. Strikingly, the measured protein levels for the SV40 and JCV chimeras versus their H42Q mutant derivatives were nearly identical, yet only the wild-type chimeras were functional in the biological assays used.
Figure 4.
Expression of DnaJ, or J-domain chimeras of the pWKG90 vector series, in strain WKG190 grown at 30°C in the presence of no arabinose inducer (lane A), or 66 μM, 660 μM, 6.6 mM, or 33 mM l-arabinose (lanes B–E, respectively). Arrows show the position of the induced proteins where only the relevant J-domain of each expression plasmid is indicated in the left margin. The right margin shows quantitated relative protein levels for lanes B–E of each panel, normalized to uninduced control and expressed in arbitrary units.
Full induction of DnaJ by arabinose resulted in protein levels 19- to 23-fold higher than DnaJ levels present in cells with only the chromosomal copy of dnaJ under nonstressed conditions as revealed by quantitative immunoblot analysis (data not shown). Finally, analysis of protein stability showed that all chimeras of the pWKG90 series DnaJ, and DnaJ259 had half-lives of greater than 20 min, the only exception being the protein produced by pWKG96 which had a half life of 5–10 min (data not shown).
For each of the viral chimeras of the pWKG90 series, two distinct protein species were always observed that corresponded to the expected chimera and a smaller band, in stoichiometric amounts, that represented either a strong internal initiation near residue 52–62, or specific proteolytic cleavage (data not shown). Internal initiation within the J-domain, or cleavage of the viral J-domain chimeras within the J-domain, may result in inactive proteins, and may partially explain the toxicity and/or reduced growth observed with certain chimeras. Two deletion derivatives of pWKG93, pWKG93Δ13 and pWKGΔ29, were constructed that removed 13 or 29 residues from the N terminus of the viral J-domain. Neither deletion derivative was active in any of the complementation assays. This result, together with the data of the H42Q mutations, shows that all complementing activity is derived from the larger of the two protein species corresponding to the expected molecular weight of the chimeras (data not shown).
DISCUSSION
Our results compellingly argue that residues comprising the SV40, JCV, or BKV N-terminal T/t common exon can function as a J-domain in a heterologous bacterial assay system. These results clearly imply that the viral J-domain may be interacting coordinately with various members of the Hsp70 family of chaperones within virus-infected cells. Another possibility is that the virus has exploited the J-domain as a scaffold and that residues essential to viral protein interactions with the host machinery are exposed along the scaffold. Of course, the two possibilities are not mutually exclusive. Several workers have reported Hsp70 association with T to be dependent upon the presence of the T N-terminal segment and that the sequences necessary for this interaction have been delimited to residues 1–97 including the J-domain (58, 59). In addition, SV40 T nuclear localization appears to be critically dependent upon cytoplasmic Hsp70 or Hsc70 (60–62).
A survey of reported mutations in the viral T/t common domain suggests that lesions located in homologous J-domain structural motifs can abolish a wide range of biological activities attributed to T. Two physically separable activities of SV40 T—(i) pRB binding to residues encompassing residues 105–114 and (ii) association of the N-terminal 1–82 residues with p300—appear necessary for T-mediated bypass of p53 cell cycle arrest at the G1 restriction point (28). T mutants mapping within putative helices 1–2 of the viral J-domain impair or abrogate this bypass of p53 cell cycle arrest. Four lesions within the N-terminal region that abolish p300-associated transforming function, but do not impair either pRB binding or p53 binding, can be complemented by the adenoviral E1A protein, thus supporting the notion that the ability to target and sequester p300 is essential for transformation (30). The pRB-like proteins, p130 and p107, undergo cell cycle-dependent phosphorylation and form complexes with transcription factor E2F-4,5 subtypes (21). Although the precise roles of p107 and p130 as anti-oncoproteins are still unclear, expression of SV40 T, or a truncated SV40 T 1–147 can apparently modulate their phosphorylation state. A mutant within the putative J-domain helix 3 does not impair T binding to p130, but prevents T-mediated alteration of its phosphorylation state, thus implying that an N-terminal region could influence cell cycle-dependent posttranslational modifications of p130 (29).
Hsp70 chaperone machines perform diverse roles, from promotion of refolding, prevention of aggregation, assembly/disassembly of protein complexes, to targeting and presentation of certain substrates destined for degradation, among others. Our results lead us to propose that some aspects of the viral life cycle are the results of viral J-domain/Hsp70 interaction, and that therefore, the T antigens might exploit one, or several, of the activities attributed to Hsp70 chaperone machines. Recent results demonstrate restrictions to DnaJ/Hsp70 pair complementation and imply a role for cognate pair recognition for proper chaperone/cochaperone interaction (63, 64) Masking the accessibility of a J-domain by protein–protein interactions, by compartmentalization, or by posttranslational modifications could further provide the framework for controlled chaperone switch mechanisms. The recognition of the viral T/t common exon as a J-domain sheds new light upon the interpretation of the phenotypes of mutations within this domain, and demonstrates the potential for control of the viral program by molecular chaperones.
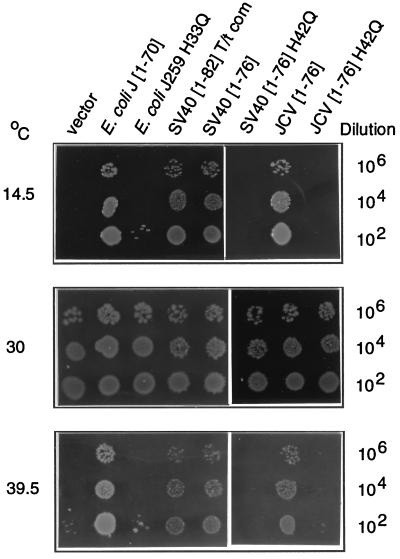
Figure 3.
Bacterial viability assay in strain WGK190 using J-domain chimeras made in the pWKG90 series of vectors (Fig. 2 and Table 1). Shown is a representative set of complementation tests for bacterial growth on plates containing 6.6 mM l-arabinose at the indicated temperatures, under conditions described in the legend to Table 1. Only the relevant J-domain is indicated at the top of the figure, as plasmids are identical to those depicted in Table 1.
Acknowledgments
We thank Luz-Maria Guzman and Dominique Belin for pBAD22, LMG190, and advice on the arabinose-inducible vector system; Chiharu Ueguchi for CU247; Joe Curran, Hans Türler, and Richard Frisque for pUC19–SV40, JCV (pMad1-TC), and BKV (pBKV-9); Kurt Wüthrich, Maurizio Pellecchia, and Thomas Szyperski for discussion and sharing NMR results prior to publication; and finally Sam Landry, Dan Wall, and members of the laboratory for discussions. This work was supported by the Swiss National Science Foundation (FN31-47283-96), the De Reuter Foundation, and the Canton of Geneva.
ABBREVIATIONS
- eop
efficiency of plating
- JCV
JC virus
- BKV
BK virus
- SV40
simian virus 40
- pRB
retinoblastoma
- T
large T antigen
- t
small t antigen
- mt
middle-T antigen
References
- 1.Caplan A J, Cyr D M, Douglas M G. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cyr D M, Langer T, Douglas M G. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 3.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 4.Silver P A, Way J C. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- 5.Cheetam M E, Jackson A P, Anderton B H. Eur J Biochem. 1994;226:99–107. doi: 10.1111/j.1432-1033.1994.tb20030.x. [DOI] [PubMed] [Google Scholar]
- 6.Jordan R, McMacken R. J Biol Chem. 1995;270:4563–4569. doi: 10.1074/jbc.270.9.4563. [DOI] [PubMed] [Google Scholar]
- 7.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyman S K, Schekman R. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 10.Tsai J, Douglas M G. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 11.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5451–5156. [PubMed] [Google Scholar]
- 12.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1995;270:2139–2144. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- 13.Cole C N. In: Virology. 3rd Ed. Fields B N, Knipe D M, editors. Philadelphia: Lippencott/Raven; 1996. pp. 1997–2026. [Google Scholar]
- 14.Frisque R J, Bream G L, Cannella M T. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah K V. In: Virology. 3rd Ed. Fields B N, Knipe D M, editors. Philadelphia: Lippencott/Raven; 1996. pp. 2027–2043. [Google Scholar]
- 16.Collins K L, Russo A A R, Tseng B Y, Kelly T J. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dornreiter I, Copeland W C, Wang T S-F. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruda M C, Zabolotny J M, Xiao J H, Davidson I, Alwine J C. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson S D, Yu X M, Mertz J E. J Virol. 1996;70:1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weisshart K, Bradley M, Weiner B M, Schneider C, Moarefi I, Fanning E, Arthur A K. J Virol. 1996;70:3509–3516. doi: 10.1128/jvi.70.6.3509-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalvide J, DeCaprio J A. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVey D, Woelker B, Tegtmeyer P. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prives C. Cell. 1990;61:735–738. doi: 10.1016/0092-8674(90)90179-i. [DOI] [PubMed] [Google Scholar]
- 24.Sontag E, Federov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 25.Dunant N M, Senften M, Ballmer-Hofer K. J Virol. 1996;70:1323–1330. doi: 10.1128/jvi.70.3.1323-1330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 27.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quartin R S, Cole C N, Pipas J M, Levine A J. J Virol. 1994;68:1334–1341. doi: 10.1128/jvi.68.3.1334-1341.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stubdal H, Zalvide J, DeCaprio J A. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaciuk P, Carter M C, Pipas J M, Moran E. Mol Cell Biol. 1991;11:2116–2124. doi: 10.1128/mcb.11.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeken M R. J Virol. 1993;67:7684–7689. doi: 10.1128/jvi.67.12.7684-7689.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W-B, Bikel I, Marsilio E, Newsome D, Livingston D M. J Virol. 1994;68:6180–6187. doi: 10.1128/jvi.68.10.6180-6187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins B S, Pipas J M. J Biol Chem. 1995;270:15377–15384. doi: 10.1074/jbc.270.25.15377. [DOI] [PubMed] [Google Scholar]
- 34.Conzen S D, Cole C N. Oncogene. 1995;11:2295–2302. [PubMed] [Google Scholar]
- 35.Marsilio E, Cheng S H, Schaffhausen B, Paucha E, Livingston D M. J Virol. 1991;65:5647–5652. doi: 10.1128/jvi.65.10.5647-5652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montano X, Millikan R, Milhaven J M, Newsome D A, Ludlow J W, Arthur A A, Fanning E, Bikel I, Livingston D M. Proc Natl Acad Sci USA. 1990;87:7448–7452. doi: 10.1073/pnas.87.19.7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peden K W C, Pipas J M. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 38.Sompayrac L, Danna K J. Virology. 1992;191:439–442. doi: 10.1016/0042-6822(92)90206-5. [DOI] [PubMed] [Google Scholar]
- 39.Spence S L, Pipas J M. Virology. 1994;204:200–209. doi: 10.1006/viro.1994.1524. [DOI] [PubMed] [Google Scholar]
- 40.Kelley W L, Landry S J. Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 41.Hill R B, Flanagan J M, Prestegard J H. Biochemistry. 1995;34:5587–5596. doi: 10.1021/bi00016a033. [DOI] [PubMed] [Google Scholar]
- 42.Pellecchia M, Szyperski T, Wall D, Georgopoulos C, Wüthrich K. J Mol Biol. 1996;260:236–250. doi: 10.1006/jmbi.1996.0395. [DOI] [PubMed] [Google Scholar]
- 43.Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wüthrich K. Proc Natl Acad Sci USA. 1994;91:11343–11347. doi: 10.1073/pnas.91.24.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueguchi C, Shiozawa T, Kakeda M, Yamada H, Mizuno T. J Bacteriol. 1995;177:3894–3896. doi: 10.1128/jb.177.13.3894-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 47.Bukau B, Walker G. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ang D, Georgopoulos C. J Bacteriol. 1989;171:2748–2755. doi: 10.1128/jb.171.5.2748-2755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ang D, Chandrasekhar G N, Zylicz M, Georgopoulos C. J Bacteriol. 1986;167:25–29. doi: 10.1128/jb.167.1.25-29.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang P J, Craig E A. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R F, Kushner S R. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 52.Kunkel T A. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 53.Liberek K, Wall D, Georgopoulos C. Proc Natl Acad Sci USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szabo A, Korszun R, Hartl F U, Flanagan J. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 55.Alfano C, McMacken R. J Biol Chem. 1989;264:10709–10718. [PubMed] [Google Scholar]
- 56.Zylicz M, Ang D, Liberek K, Georgopoulos C. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westermann B, Gaume B, Herrmann J M, Neupert W, Schwarz E. Mol Cell Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May E, Breugnot C, Duthu A, May P. Virology. 1991;180:285–293. doi: 10.1016/0042-6822(91)90033-8. [DOI] [PubMed] [Google Scholar]
- 59.Sawai E T, Rasmussen G, Butel J S. Virus Res. 1994;31:367–378. doi: 10.1016/0168-1702(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 60.Imamoto N, Matsuoka Y, Kurihara T, Kohno K, Miyagi M, Sakiyama F, Okada Y, Tsunasawa S, Yoneda Y. J Cell Biol. 1992;119:1047–1061. doi: 10.1083/jcb.119.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y, Thomas J O. Mol Cell Biol. 1992;12:2186–2192. doi: 10.1128/mcb.12.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, DeFranco D B. Mol Cell Biol. 1994;14:5088–5098. doi: 10.1128/mcb.14.8.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cyr D M, Douglas M G. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- 64.Schlenstedt G, Harris S, Risse B, Lill R, Silver P A. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]