Abstract
The SH3 domains of src and other nonreceptor tyrosine kinases have been shown to associate with the motif PXXP, where P and X stand for proline and an unspecified amino acid, but a motif that binds to the SH3 domain of myosin has thus far not been characterized. We previously showed that the SH3 domain of Acanthamoeba myosin-IC interacts with the protein Acan125. We now report that the Acan125 protein sequence contains two tandem consensus PXXP motifs near the C terminus. To test for binding, we expressed a polypeptide, AD3p, which includes 344 residues of native C-terminal sequence and a mutant polypeptide, AD3Δ977–994p, which lacks the sequence RPKPVPPPRGAKPAPPPR containing both PXXP motifs. The SH3 domain of Acanthamoeba myosin-IC bound AD3p and not AD3Δ977–994p, showing that the PXXP motifs are required for SH3 binding. The sequence of Acan125 is related overall to a protein of unknown function coded by Caenorhabditis elegans gene K07G5.1. The K07G5.1 gene product contains a proline-rich segment similar to the SH3 binding motif found in Acan125. The aligned sequences show considerable conservation of leucines and other hydrophobic residues, including the spacing of these residues, which matches a motif for leucine-rich repeats (LRRs). LRR domains have been demonstrated to be sites for ligand binding. Having an LRR domain and an SH3-binding domain, Acan125 and the C. elegans homologue define a novel family of bifunctional binding proteins.
Keywords: actin, cytoskeleton, Acanthamoeba, microfilament, motor protein
Acan125, the first nonactin protein found to bind molecular myosin-I, is a soluble protein from Acanthamoeba that binds to myosin-I’s SH3 domain (1). SH3 domains are present in the sequences of myosin-Is from a variety of organisms, including humans (2–4). To date, only the SH3 domain of Acanthamoeba myosin-I has a candidate ligand—i.e., Acan125. In contrast, binding proteins have been identified for SH3 domains of src and a host of nonreceptor tyrosine kinases and adapter proteins (5). These proteins are intimately involved in signal transduction, raising the possibility that myosin-I might also be linked through SH3 to a signaling pathway.
Myosin-I has two domains, an N-terminally located motor domain and a C-terminal tail. The head domain shares considerable sequence homology with other myosins, but the tail is quite divergent. The tail functions in part to attach myosin-I to a substratum, which can be either actin filaments or a membrane surface (2). Membranes containing acidic phospholipids have been shown to interact directly with a region of the tail apart from the SH3 domain (6). However, phospholipid binding alone seems inadequate to explain the restricted distribution of myosin-I in cells (2), which supports the idea of a protein that targets myosin-I in the membrane. Acan125 is a candidate for localizing myosin-I in the membrane, owing to the interaction between Acan125 and myosin-I’s SH3 domain.
All known SH3-binding proteins contain the consensus sequence PXXP, where P is proline and X is a nonconserved residue (5). This consensus was determined for a large number of SH3-binding peptides representing core binding sites of the known SH3-binding proteins. The core sequence consists of seven residues, including multiple proline residues required to form a left-handed type II proline helix (7). The helix aligns prolines that comprise the PXXP sequence and arginine and lysine residues that flank the PXXP sequence to make specific contacts in the SH3 domain (7). Residues of the helix determine both the affinity of the interaction (8) and the orientation of the interaction within the binding site (9, 10). The sequence of Acan125 is expected to contain a PXXP motif that interacts with the SH3 domain of myosin-I.
To identify domains of interaction with myosin-I and possibly with other proteins, we have determined the primary structure of Acan125. The sequence contains two tandem PXXP motifs; at least one PXXP motif is shown to be required for association with the SH3 domain of myosin-I. In addition, the Acan125 sequence contains tandem repeats that conform to a motif known as a leucine-rich repeat (LRR). LRRs exhibit remarkable versatility, forming domains both extracellularly and intracellularly (11). Many of the cytoplasmically oriented LRR domains are present in signal transduction proteins (12) and often contain multiple distinct domains as exemplified by Drosophila Flightless, which has gelsolin and LRR domains (13). Acan125 is unique in combining LRR and PXXP domains and suggests intriguing functional possibilities for this protein.
EXPERIMENTAL PROCEDURES
Purification of Acan125.
Acan125 was purified from Acanthamoeba castellanii Neff strain grown in liquid culture. Cells were harvested, washed free of culture medium, and resuspended 1:2 (wt/vol) in TBS (0.15 M NaCl/0.006 M KCl/0.05 M Tris·HCl, pH 8.0) containing 10 μg/ml leupeptin, 1 μg/ml pepstatin A, 0.10 mM phenylmethylsulfonyl fluoride, and 10 units/ml aprotinin. Cells were broken with a Dounce homogenizer and the lysate was clarified by centrifugation at 100,000 × g. The supernatant was fractionated with solid ammonium sulfate. Proteins that precipitated between 22% and 55% of saturation were dissolved with 1.0 M ammonium sulfate and applied to phenyl-Sepharose 6B; the column was eluted in a single step by removal of ammonium sulfate. The eluate was fractionated on an affinity column; glutathione beads were coupled with a fusion protein of glutathione S-transferase (GST) and the SH3 domain of Acanthamoeba myosin-IC (1). The fraction that eluted with 5× TBS was loaded onto mono-Q (Pharmacia) equilibrated with 25 mM 3-(N-morpholino)propanesulfonic acid, pH 7.0/50 mM KCl/0.2 mM dithiothreitol. A linear gradient to 500 mM KCl eluted Acan125, which was identified by Western blotting with Acan125 antibodies as a single band in a Coomassie blue-stained gel after SDS/PAGE. We obtained 200 μg of purified protein from 230 g of cells.
Protein Microsequencing.
Peptide sequence was obtained from Acan125 immobilized on poly(vinylidene difluoride) (PVDF). Acan125 partially purified as described previously (1) was separated by SDS/PAGE and transferred to PVDF. The band, shown previously to be reactive with Acan125 antibodies, was excised; the protein on the membrane was digested with endoproteinase LysC (Protein Sequencing Core, University of Texas Southwestern Medical Center at Dallas). The peptides were separated by reversed-phase HPLC. Several peaks were collected and applied directly to an Applied Biosystems sequencer.
Peptide sequence was also obtained from column-purified Acan125. Purified Acan125 (150 μg) was reconstituted in 600 μl of 0.10 M Tris·HCl, pH 9.25, and 10% (vol/vol) acetonitrile to which was added 3 mg of endoproteinase LysC (Wako). The mixture was incubated at 37°C overnight and then made 2% trifluoroacetic acid prior to reversed-phase HPLC on a 2.1 × 250 mm C18 reversed-phase column (Vydac 218TP). The peptides were chromatographed using a gradient of 0–40% (vol/vol) acetonitrile in 0.1% trifluoroacetic acid over 60 min followed by 40–80% acetonitrile in 0.1% trifluoroacetic acid over 30 min at a flow rate of 100 ml/min; peaks were automatically collected by using a Pharmacia SMART liquid chromatography system. Peaks judged homogeneous by UV absorbance were applied directly to a Hewlett Packard G1000A protein sequencer. Other peaks were subject to rechromatography in the same chromatography system, using shallower gradients prior to sequencing.
Library Screening.
cDNA libraries in λZap II phage vector (Stratagene) were screened with antibodies to Acan125. Escherichia coli XL1-Blue MRF′ cells were infected with approximately 3 × 105 phages and grown on 10 × 150 mm plates for 3.5–4 hr at 42°C. Plaques were transferred to 10 mM isopropyl β-d-thiogalactoside-immersed nitrocellulose membranes (DuPont/NEN) for 2 hr at 37°C. The membranes were lifted, blocked, and then probed sequentially with blot-purified rabbit anti-Acan125 antibodies and horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies. Positive plaques were visualized by autoradiography using ECL reagent (Amersham). Phage were purified and rescued by in vivo excision to pBluescript phagemid using the supplier’s protocol (Stratagene). A positive clone, AD3, was identified as an 1186-bp insert that codes for the 3′ end of the Acan125 message.
5′ Rapid Amplification of cDNA Ends (RACE).
The 5′ end of AD3 was extended on a cDNA template synthesized from Acanthamoeba RNA. First-strand cDNA was synthesized from 5 μg of total Acanthamoeba RNA primed with 5′-TTAAGCTTCTCCACCGCGCTCTTC-3′ using Superscript II reverse transcriptase (Life Technologies). Synthetic oligonucleotides for PCR included the antisense primer 5′-CTCGGATCCCTTGTATTCCTTGACC-3′, which was based on AD3 sequence, and the sense primer 5′-CTCCACTTCAACAACTACTTCATCGG-3′, which was synthesized on the basis of the peptide sequence LHFNNYFIG. With the first strand cDNA as a template, the primers produced a single product of 1706 bp by PCR using Expand high-fidelity Taq DNA polymerase (Boehringer Mannheim). The final PCR product was ligated into pBluescript (Stratagene) by the T-A cloning method (14) and used to transform E. coli DH5α. The resulting clone was designated S7-AD3AS3.
The end of the coding region was obtained by 5′ RACE extension of S7-AD3AS3 cDNA. An antisense primer based on S7-AD3AS3 cDNA, 5′-CTTCTCGTTCTTGAGCAGGTC-3′, was used to synthesize the first-strand cDNA as described above, and the 3′ end was tailed with dTTP using terminal deoxynucleotidyltransferase (Boehringer Mannheim). The resulting cDNA was primed with an antisense primer, 5′-CTTCATGGAGTCCTTGTCCAGG-3′, and a 1:19 mixture of sense primers, 5′-GACTCGAGTCGACATCGA(T)17-3′ and 5′-GACTCGAGTCGACATCGA-3′, and amplified by PCR. A DNA fragment of 859 bp was generated and inserted into pBluescript.
Expression of AD3 and AD3Δ977–994.
AD3Δ977–994 was prepared from AD3 that had been subcloned into pGex-2T-KG (15) by site-directed mutagenesis performed using a Transformer kit (CLONTECH) according to the manufacturer’s protocol. A mutagenic primer, 5′-CAGTCGGCCACCGACACGGCCAAGCCGCTG-3′, was prepared such that bases corresponding to 2914–2928 and 2983–2997 of Acan125 are joined. A clone with the desired mutation was identified by restriction digestion and verified by double-stranded DNA sequencing. This procedure resulted in an in-frame deletion of 54 bp corresponding to residues 977–994 of Acan125 sequence, which contain both PXXP motifs.
The polypeptides AD3p and AD3Δ977–994p were expressed as fusion proteins with hexahistidine-tagged thioredoxin (Trx). To create the vector, the Trx open reading frame of pTrxFus (Invitrogen) was cut out with NdeI and BamHI and subcloned into pET28b (Novagen) cut with the same enzymes. The resulting plasmid, pET28b-Trx, expresses an open reading frame with hexahistidine fused to the N terminus of Trx under control of the T7 lac promotor. AD3 and AD3Δ977–994 inserts were cut out of pBluescript SK(−) with EcoRI and XhoI and subcloned into the same sites of pET28b-Trx. Recombinant plasmids were used to transform E. coli BL21 (DE3, pLysS). Cultures (4 ml) were grown in Terrific Broth (16) containing 60 μg/ml kanamycin at 35°C; synthesis of fusion protein, Trx-AD3p or Trx-AD3Δ977–994p, was induced by the addition of 1 mM isopropyl β-d-thiogalactoside at room temperature for 5 hr. Hexahistidine-tagged protein in a lysate of sonicated cells was isolated with 0.5 ml of nickel chelated by nitrilotriacetic acid resin according to manufacturer’s protocol (Qiagen). The purification procedure resulted in the recovery of 20 μg of Trx-AD3Δ977–994p and 40 μg of Trx-AD3p.
In Vitro Binding Assays.
Tests for the binding of Trx fusion proteins were performed with GST-SH3 immobilized on beads. The SH3 domain of Acanthamoeba myosin-IC, expressed as fusion protein with GST, and GST alone were coupled to glutathione beads (1). Approximately 15 ml of packed beads was combined with purified Trx fusion protein in a total volume of 250 ml of 16 mM NaH2PO4, pH 8.0/96 mM NaCl/80 mM imidazole and incubated 20 min at 4°C. The beads were recovered by centrifugation and washed; protein in the supernatant was precipitated by addition of 5 vol of acetone and centrifuged, and the pellet was air dried. Pellets and beads were heated in 40 μl of sample buffer; proteins in 15 μl of solubilized samples were separated by SDS/PAGE.
SDS/PAGE.
SDS/PAGE and Western blotting were performed as described previously (1). Proteins in the gel were stained with Coomassie blue R-250. Densitometry was performed using packaged software (Molecular Analyst, Bio-Rad).
RESULTS AND DISCUSSION
Cloning and Sequencing of Acan125.
To identify relevant clones, we determined amino acid sequences of peptides isolated from Acan125 protein by using two approaches. First, Acan125 was partially purified from a lysate by association with the SH3 domain of Acanthamoeba myosin-IC immobilized on beads and isolated as a single band in SDS/PAGE. The band was transferred to PVDF and sequenced directly. Several blank cycles were obtained, indicating that the N terminus is blocked. Subsequently, the band of Acan125 on PVDF was digested and the digest was separated by HPLC; 6 peptides were isolated and sequenced. Second, Acan125 was purified to homogeneity and digested. The digest was separated by HPLC and 16 peptides were isolated and sequenced. Both approaches utilized an Acanthamoeba myosin-IC SH3 affinity column, reactivity with antibodies to Acan125, and digestion with endoproteinase LysC. Of the 22 peptides, 4 were found by both approaches, indicating that all peptides were derived from the same protein.
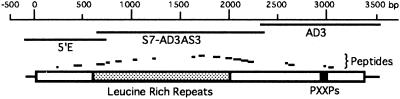
We identified a clone containing Acan125 peptide sequence from a screen of an expression library with polyclonal antibodies to Acan125. The clone, AD3, measuring 1186 bp, has an open reading frame containing four peptide sequences (Fig. 1). In addition to coding sequence, AD3 contains a stop codon, untranslated sequence, and a polyadenylylation signal, indicating the 3′ end of the Acan125 message.
Figure 1.
Scale drawing summarizing the results of cloning and sequencing of Acan125. Clone AD3 was obtained by screening an expression library, and clones S7-AD3AS3 and 5′E were obtained by 5′ RACE reactions. The peptide sequences were obtained by direct sequencing of the Acan125 protein purified by either column chromatography or SDS/PAGE; four peptides were obtained by both methods. The nucleotide and protein sequences can be retrieved from the GenBank database (accession no. U89688U89688). The block diagram depicts the domain organization of Acan125 as described in the text.
The 5′ end of Acan125 was obtained by consecutive RACE reactions using cDNA synthesized from Acanthamoeba RNA as a template. A primer in the AD3 clone and an upstream primer based on a peptide sequence yielded a 1706-bp PCR product. The translated sequence contains 12 peptide sequences (Fig. 1). Subsequently, a primer based on the 1706-bp sequence and a primer complementary to the presumed 5′ end of the mRNA generated an 859-bp PCR product. An open reading frame was identified that contained the remaining 2 peptides. Upstream of the reading frame is a stop codon, which marks the 5′ untranslated sequence. Between the stop codon and the peptide sequence, two ATG codons are found to be in frame. The sequence surrounding neither ATG contains the consensus for start codons described by Kozak (17). We surveyed the GenBank database and found that 8 of 10 Acanthamoeba nucleotide sequences do not conform to the Kozak consensus. To determine the probable start codon, we used the aligned sequences of Acan125 and a homologous gene product from Caenorhabditis elegans (see below). A contiguous group of three identical residues in the aligned sequences was found between the potential start codons. Therefore, we conclude that the first ATG after the stop codon codes for the initiator methionine.
The merged cDNA sequence codes for a protein with 1121 amino acids and calculated Mr 121,608. The sequences of the three clones, 1186 bp (nucleotides 2332–3518), 1706 bp (nucleotides 661-2367), and 859 bp (nucleotides −95 to 729 plus 35 bp of RACE primer), were shown to overlap. All of the peptides microsequenced were accounted for in the deduced sequence, suggesting a single isoform. The sequence codes for an Acan125-reactive protein, since antibodies to Acan125 recognized both the protein used to generate peptide sequence and protein expressed by AD3.
Of the total number of bases sequenced using Taq, 1267 in the coding region were not verified by overlapping clones and peptides. Assuming that fidelity errors are Poisson distributed, the probability of having no errors is 0.88 and the probability of having a single base error is 0.11. This is based on the error rate of the Taq DNA polymerase preparation used (8.5 × 10−6, Boehringer technical information) and 25 cycles of amplification, giving a mutation rate of 1 × 10−4 (18). The probability of Taq having introduced a single error in the amino acid sequence is less than 0.11 owing to codon degeneracy. The possibility of an error leading to a frameshift is remote, given the large number of peptide sequences used to verify the correct clones.
Identification of SH3 Binding Domain.
The SH3 binding domain was localized in the native sequence by partial digestion of Acan125. The native Acan125 protein was found to be highly susceptible to endogenous proteolysis during purification, yielding a stable fragment of ≈110 kDa. This fragment was found not to bind immobilized GST-SH3 (data not shown), indicating that an SH3-binding domain resides ≈15 kDa from one end of Acan125. At the N terminus, this corresponds to a region of Acan125 which precedes the LRR domain (Fig. 1; see below) and which does not contain a PXXP sequence. In contrast, the C-terminal 25% of Acan125 is rich in prolines with a number of PXXP sequences. Thus, the C terminus of Acan125 most likely contains the functional SH3-binding domain.
Though a number of PXXP sequences appear in Acan125, only two of the PXXP sequences conform closely to the consensus PXXP motif for peptides that bind to previously characterized SH3 domains (Table 1). As a group, these peptides bind to an SH3 domain stereospecifically in the minus orientation; a separate group of peptides bind to the same SH3 domain in the plus orientation (9, 10). It is striking that the arginine at the P−3 position is conserved in the Acanthamoeba sequences (Table 1). No peptide in the Acan125 sequence conforms well to the consensus for peptides that bind an SH3 domain in the plus orientation. The two PXXP motifs of Acan125, separated by only two residues, are present within the proline-rich region ≈15 kDa from the C terminus of Acan125, suggesting one or both motifs associate with the SH3 domain of myosin-I.
Table 1.
Sequence alignments of PXXP motifs
| PXXP motif | P3 | P2 | P1 | P0 | P−1 | P−2 | P−3 |
|---|---|---|---|---|---|---|---|
| Synthetic library consensus for Src | X | P | P | L | P | X | R |
| Synthetic library consensus for Pl3K | X | P | P | L | P | X | R |
| Human dynamin (812–818) | A | P | P | V | P | S | R |
| Human p47phox (362–368) | Q | P | A | V | P | P | R |
| Murine SOS1 (1153–1159) | P | P | P | V | P | P | R |
| Murine SOS1 (1214–1220) | P | P | L | L | P | P | R |
| Murine SOS1 (1292–1298) | G | P | P | V | P | P | R |
| Acanthamoeba Acan125 (979–985) | K | P | V | P | P | P | R |
| Acanthamoeba Acan125 (988–994) | K | P | A | P | P | P | R |
| C. elegans K07G5.1p (935–941) | P | P | P | L | P | Q | R |
| C. elegans K07G5.1p (951–957) | P | P | L | L | P | P | K |
| C. elegans K07G5.1p (969–975) | S | P | T | T | P | P | P |
| Consensus sequence | X | P | p | Ψ | P | p | r |
The tabulated PXXP motifs are peptide sequences that bind SH3 in the minus orientation (7). P3 to P−3 are sites in the SH3 domain that bind residues of the ligand (9, 10). X and Ψ represent nonconserved and hydrophobic residues. Lowercase letters indicate that the residue is conserved in at least 50% of the sequence.
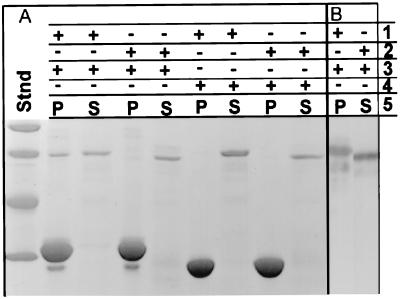
To test for SH3 binding, we expressed the clone AD3 (Fig. 1), which codes for a major segment of the Acan125 protein (≈39 kDa) containing the entire proline rich region. We expressed AD3p, containing native Acan125 sequence, and AD3Δ977–994p, lacking the sequence, RPKPVPPPRGAKPAPPPR, including both putative PXXP motifs, as fusion proteins with Trx. We expressed the SH3 domain of Acanthamoeba myosin-IC as a fusion protein with GST. The binding assay included an excess of GST or GST-SH3 immobilized on glutathione beads (3–5 mg total each) with respect to Trx-AD3p and Trx-AD3Δ977–994p (Fig. 2A). Beads containing GST-SH3 sedimented ≈46% of the total Trx-AD3p and less than 1% of the total Trx-AD3Δ977–994p, which were each present at ≈300 nM. Given near half-maximal binding at ≈300 nM AD3p, the data imply a submicromolar affinity, which compares favorably with the micromolar affinities reported for synthetic peptide binding to the SH3 domains of src and abl (8). The binding of Trx-AD3p is not mediated by the GST moiety, since GST coupled to beads did not sediment either Trx-AD3p or Trx-AD3Δ977–994p (Fig. 2A). Thus, only the PXXP motifs present between residues 977 and 994 are shown to be required for optimal binding of AD3p to the SH3 domain of myosin-I.
Figure 2.
Identification of a binding site for the SH3 domain of Acanthamoeba myosin-IC in the sequence of Acan125 using an affinity ligand approach. (A) Uncut gel stained with Coomassie blue. (B) Two lanes of a Western blot using anti-Acan125 antibodies. Lines 1–4 indicate the presence or absence of reactants in the binding assay: 1, Trx-AD3; 2, Trx-AD3Δ977–994; 3, GST-SH3; and 4, GST. Line 5 refers to the origin of the samples: either pellet (P) containing proteins associated directly or indirectly with glutathione-Sepharose beads or supernatant (S) containing proteins not associated with the beads. GST (0.32 mg/mg of beads) and GST-SH3 (0.35 mg/mg of beads) were coupled directly to glutathione-Sepharose beads; note that GST (≈27 kDa) and GST-AD3 (≈32 kDa) sediment quantitatively. Also present in the binding assay as indicated were (final concentrations) 0.016 mg/ml Trx-AD3 (A and B), 0.016 mg/ml Trx-AD3Δ977–994 (A), or 0.008 mg/ml Trx-AD3Δ977–994 (B). Note that anti-Acan125 antibodies reacted with Trx-AD3 (≈65 kDa) and Trx-AD3Δ77–994 (<≈65 kDa) but not GST-SH3 in the pellet (B).
Bands corresponding to Trx-AD3p and Trx-AD3Δ977–994p react with antibodies to Acan125 (Fig. 2B). Trx-AD3p and Trx-AD3Δ977–994p with calculated masses of ≈53 kDa appear to be ≈22% larger on SDS/PAGE (Fig. 2A). The fact that native Acan125 migrates normally may be related to its overall size, since we find a GST fusion protein of AD3p (and AD3Δ977–994p) to have an apparent mass by SDS/PAGE only 7–8% above its calculated mass of ≈65 kDa (data not shown). The fusion proteins may be retarded by the clusters of proline and charged residues at the C terminus, as has been described (19–21).
The possibility that we have identified an SH3 binding domain in native Acan125 rests on whether a linear sequence likely binds to SH3. A linear sequence, the core binding sequence, is known to interact directly with a fold in the SH3 domain, and sites of interaction within the binding fold have been mapped using peptide (7) and native protein (22) ligands. The structural data support our conclusion that one or both PXXP motifs in the sequence 977–994 of Acan125 directly interact with the SH3 domain of myosin-I.
C. elegans Homologue.
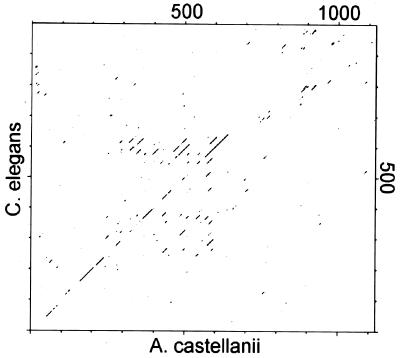
The Acan125 protein sequence was found in a blast (basic local alignment search tool) search to be similar to an uncharacterized gene from C. elegans, K07G5.1 (GenBank accession no. Z71264Z71264). Alignment of the sequences using the GCG program BestFit shows the two sequences to have 48% similarity and 25% identity overall. Relatedness in the aligned sequences is confirmed by a dot matrix analysis showing a diagonal line (Fig. 3). On the basis of this analysis, the Acanthamoeba and C. elegans proteins exhibit signs of homology over the first two-thirds of the sequence, ending with residue ≈640 (Fig. 3). Thus, C. elegans gene K07G5.1 appears to code for an Acan125-like-protein (ALP).
Figure 3.
Dot plot illustrates the regions of similarity between the protein sequence of Acan125 and the translation of C. elegans gene K07G5.1. The sequences were aligned with the GCG program compare, using a window of 25 and a stringency of 14.
Despite the lack of related sequence, the C-terminal portions of the Acanthamoeba and C. elegans proteins do share potential SH3-binding sites. Three potential tandem PXXP motifs in the C. elegans ALP sequence align with the consensus motif for SH3-binding peptides (Table 1). Two of the C. elegans ALP motifs conform to the consensus as well as the Acan125 motifs (Table 1); a third motif (969–975, Table 1) appears less well conserved. The presence of PXXP motifs suggests that corresponding nonhomologous regions of Acan125 and C. elegans ALP may be functionally related.
Identification of LRRs.
A blast search using a filter for low complexity and repetitive sequences demonstrates that segments of Acan125 likely contain LRRs. An LRR, as revealed in the structure of ribonuclease/angiogenin inhibitor protein, RAI, is a single strand of β-sheet and α-helix (23). An individual LRR of RAI is 28–29 residues and has the form XLXXLXLXXN/CXLX7LXXXLX4, where the subscript denotes the number of repeating residues. The residues of the β-strand and adjacent loops (underlined) are considered the most invariant. Other LRR proteins substitute more liberally for conserved residues than RAI, which contains uniquely regular repeats. Variability is reflected in the consensus sequence for the superfamily of LRRs, which has the form XLXXLXLXXNX1–2aX2–3aX0–4aX0–2aX1–5, where L is given if leucine is present in at least 50% of the repeats and a is given if A, V, L, I, F, Y, or M is present in at least 80% of the repeats (11). Cysteine and threonine are the most common substitutions found for asparagine at position 10. The length of the helix (nonunderlined residues) varies among members of the superfamily (11, 12).
We identified 16 repeats in the central region of the sequences of Acan125 and C. elegans ALP (Table 2). The consensus sequence (Table 2) matches the LRR consensus save for the overall length of some repeats. Acan125 and C. elegans ALP exceed the typical LRR length of 29 in 12 of 32 repeats. The additional length is found exclusively in the putative helical domain, which is not as well conserved as the region surrounding the β-strand. Acan125 and C. elegans ALP repeats do substitute for leucine at position 7 slightly more than average, but so do a number of other families of LRR proteins (11). Therefore, we propose Acan125 and C. elegans ALP belong to the superfamily of LRR proteins, having a single domain of at least 16 repeats.
Table 2.
Sequence alignments of LRRs
| LRR | Species | Sequence | C-terminal residue |
|---|---|---|---|
| 1 | A.c. | HVKKFNMKEFEQPVTPNELKA-------LVDTLHFNN | 224 |
| C.e. | KLRDLKIDDFSH-LLPKDLLP-------IVGVLQYSA | 221 | |
| 2 | A.c. | YFIGF-VARDYRLDKDSMK--------VLTDMIGVNT | 253 |
| C.e. | YFTGL-ICDAVRVSSEVID--------VVLSVIRKSQ | 250 | |
| 3 | A.c. | KMEELVLSRVEGTASAFE---------ELGQAIAKNRNS | 283 |
| C.e. | NLKKLQLRSCSLPKDFIT---------LLASAFHNNQNA | 280 | |
| 4 | A.c. | ALTSIDWSNNLIKDAGVA---------ALAAAVASMGH | 312 |
| C.e. | CLEYLDLSRNPIDDKKGFT--------ILSSVLPRLN | 309 | |
| 5 | A.c. | GLTSISLKGGDATKKGTVALCTA-----FKKNVEMSR | 344 |
| C.e. | QLKYVNFSECQLSDKSINLLCTG-----LYNGMTSCKSGGM | 345 | |
| 6 | A.c. | TLTVLNLAGNRLDSDGTSA---------LAAFVSGPN | 372 |
| C.e. | QLTELILSANPMK-DDISG---------IINLVSIST | 372 | |
| 7 | A.c. | ALQTLNISGTAANLE------------MLLPAVMRGCT | 398 |
| C.e. | SLRVLDFSDTGIHLD------------KLWNSLKFGGL | 398 | |
| 8 | A.c. | ELEKFNISHNKVTAKTGP---------ELKKFLQSCG | 426 |
| C.e. | QIEKLILNGCSLSKKSEGVQ-------TAKEYFSMAV | 428 | |
| 9 | A.c. | RLSELHMRDTAVPVQVVRD--------VIKAIIGNNFIT | 458 |
| C.e. | NLSHISFNNTSMPSEYLKA--------VLLGLASNQQLQ | 460 | |
| 10 | A.c. | DF-QLDLAANKLGVLGAN---------MLAG-LAAEIT | 484 |
| C.e. | PF-RLDLDAT-CEKGSAS---------VLDACIGGI | 484 | |
| 11 | A.c. | TIKSLDLTDNDFGDEGMS---------IIADGLCHNS | 512 |
| C.e. | RCETLSLRDNNLDGELQG---------VLQSLM-MVT | 511 | |
| 12 | A.c. | SLRELHLGDN-WTRNKTKARSQAVDN--LIELI---SSEC | 546 |
| C.e. | CLRRLDIGGSNMNQLKKNNKQVHVINKVLLDVVKLYSEEG | 551 | |
| 13 | A.c. | PLHKLDLS-CKVADNQIKT--------DILPFIYSLATND | 577 |
| C.e. | CLEELNLSDCRLGAYLS----------VLINTL--AATT | 578 | |
| 14 | A.c. | TLKELDISGNAMGDKVGI---------ALGKAFQINR | 605 |
| C.e. | TLKSLDISGNEMGNFGAR---------ILSKALQVNV | 606 | |
| 15 | A.c. | TLSSLIWDRNGTRYAGFLG---------LKNGLERNN | 633 |
| C.e. | SLRSVSIDNNHIGADGFVD---------LATSIKMNH | 634 | |
| 16 | A.c. | VLVNMPVPVFDISD-LLKNEKPE----SAL-QVQQ | 662 |
| C.e. | TLTHFPYPVHDAFDCMQRVERPR----TVA-ALSQ | 664 | |
| Consensus | XLXXLXaXXnXaXXXXXXX±±±±±±±±aLaXaaXXXX±±±± | ||
Preliminary alignment of Acan125 protein and C. elegans K07G5.1 gene product sequences by GCG BestFit were refined by inspection. The species are Acanthamoeba castellanii (A.c.) and Caenorhabditis elegans (C.e.). The C-terminal residue refers to the C terminus of the sequence shown, numbered from the initiator methionine. Letters in boldface indicate consensus residues. The consensus includes an L when >50% of the sequences contain leucine, an a when >80% of the sequences contain A, V, L, F, Y, or M, an n when >80% of the sequences contain N, C, or T, an X for an unspecified amino acid, and the symbol ± for an X that is variably present.
The alignment of LRRs indicates that a more fundamental relationship exists between Acan125 and C. elegans ALP than simply the alignment of the linear sequences. Most individual LRRs of Acan125 display characteristics similar to corresponding LRRs of C. elegans ALP, including length and fidelity to the consensus motif. For example, LRRs 1, 2, 7, 10, 12, and 16 of both Acan125 and C. elegans ALP display similar extremes in insertion, in deletion, or in substitution of sequence (Table 2). Most LRR domains have one or more imperfect repeats with substitutions or deletions at critical positions in the motif (11). Whether variations in LRR structure reflect divergence is unclear, but the empirical data suggest a pressure to conserve the structure of LRRs within a family (11). On the basis of the conservation of LRR structure, Acan125 and C. elegans ALP are proposed to be members of a novel family of LRR proteins.
Of the proteins found in the blast search with Acan125, including, in order of relatedness, C. elegans K07G5, RAI, T-complex-associated-testes-expressed-1 protein, tropomodulin, and Ran GTPase-activating protein (RNA1), all but tropomodulin have previously been reported to have LRRs. The search produced a single blast hit with tropomodulin, showing that residues 219–287 of tropomodulin align with residues 572–640 of Acan125. Sequence related to the tropomodulin blast hit was found in all of the proteins above, suggesting that this segment may be derived from a common ancestral LRR gene. The LRRs of the proteins related to Acan125 share a longer motif of ≈28 residues rather than a shorter motif of ≈24 residues found in the majority of other LRR-containing proteins (24). The longer motif may have a functional significance for members of this group.
Domain Organization and Conclusion.
The family of Acan125 proteins is shown here to have at least two functional domains, LRR and PXXP (Fig. 1). The PXXP and LRR domains enable simultaneous binding of myosin-I and a ligand yet to be determined. Depending on the nature of the unknown ligand, the LRR domain could target myosin-I to the membrane, link myosin-I with another cellular process, or provide myosin-I with a novel property.
Acknowledgments
We thank Drs. Erik Bateman and Simon Atkinson (while a member of the laboratory of Tom Pollard) for supplying Acanthamoeba expression libraries and Dr. Helen Yin for supplying pGex-2T-KG. We also thank Drs. Helen Yin, Anita Zot, and Joe Albanesi for critical reading of the manuscript. This work was supported by National Science Foundation Grants MCB-9205344 and MCB-9514248 (H.G.Z.).
ABBREVIATIONS
- LRR
leucine-rich repeat
- GST
glutathione S-transferase
- PVDF
poly(vinylidene difluoride)
- RACE
rapid amplification of cDNA ends
- Trx
thioredoxin
- blast
basic local alignment search tool
- ALP
Acan125-like-protein
- RAI
ribonuclease/angiogenin inhibitor protein
Footnotes
References
- 1.Xu P, Zot A S, Zot H G. J Biol Chem. 1995;270:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- 2.Pollard T D, Doberstein S K, Zot H G. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- 3.Sellers J, Goodson H V. Prot Profile. 1995;2:1323–1423. [PubMed] [Google Scholar]
- 4.Hasson T, Mooseker M S. J Biol Chem. 1996;271:16431–16434. doi: 10.1074/jbc.271.28.16431. [DOI] [PubMed] [Google Scholar]
- 5.Feller S M, Ren R, Hanafusa H, Baltimore D. Trends Biochem Sci. 1994;19:453–458. doi: 10.1016/0968-0004(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 6.Adams R A, Pollard T D. Nature (London) 1989;340:565–588. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 8.Rickles R J, Botfield M C, Weng Z, Taylor J A, Green O M, Brugge J S, Zoller M J. EMBO J. 1994;13:5598–5604. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim W A, Richards F M, Fox R O. Nature (London) 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 10.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 11.Kobe B, Deisenhofer J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 12.Kobe B, Deisenhofer J. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 13.Claudianos C, Campbell H D. Mol Biol Evol. 1995;12:405–414. doi: 10.1093/oxfordjournals.molbev.a040215. [DOI] [PubMed] [Google Scholar]
- 14.Holton T A, Graham M W. Nucleic Acids Res. 1991;19:1156. doi: 10.1093/nar/19.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan K, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. A.2. [Google Scholar]
- 17.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelfand D H, White T J. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. New York: Academic; 1990. pp. 129–141. [Google Scholar]
- 19.Graceffa P, Jancso A, Mabuchi K. Arch Biochem Biophys. 1992;297:46–51. doi: 10.1016/0003-9861(92)90639-e. [DOI] [PubMed] [Google Scholar]
- 20.Hu C C, Ghabrial S A. J Virol Methods. 1995;55:367–379. doi: 10.1016/0166-0934(95)00085-1. [DOI] [PubMed] [Google Scholar]
- 21.John M E, Keller G. Plant Physiol. 1995;108:669–676. doi: 10.1104/pp.108.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 23.Kobe B, Deisenhofer J. Nature (London) 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 24.Kajava A V, Vassart G, Wodak S J. Structure. 1995;3:867–877. doi: 10.1016/S0969-2126(01)00222-2. [DOI] [PubMed] [Google Scholar]