Abstract
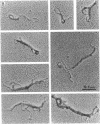
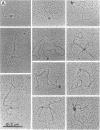
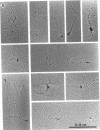
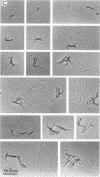
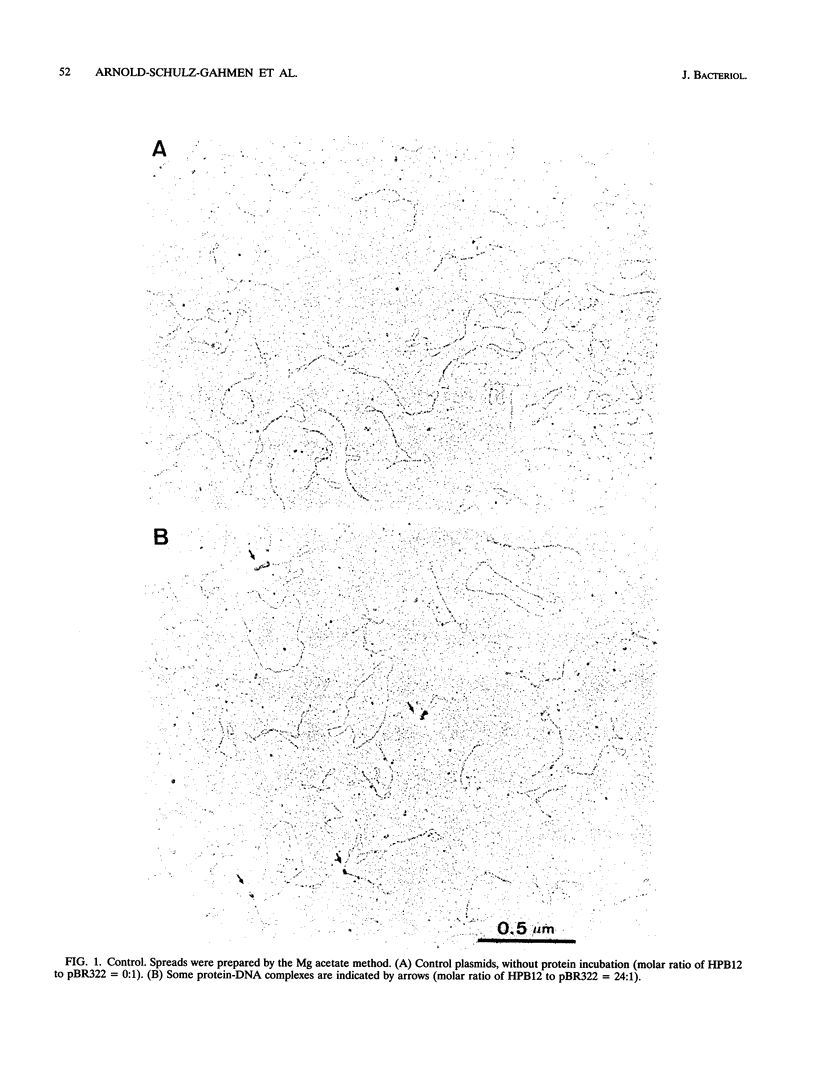
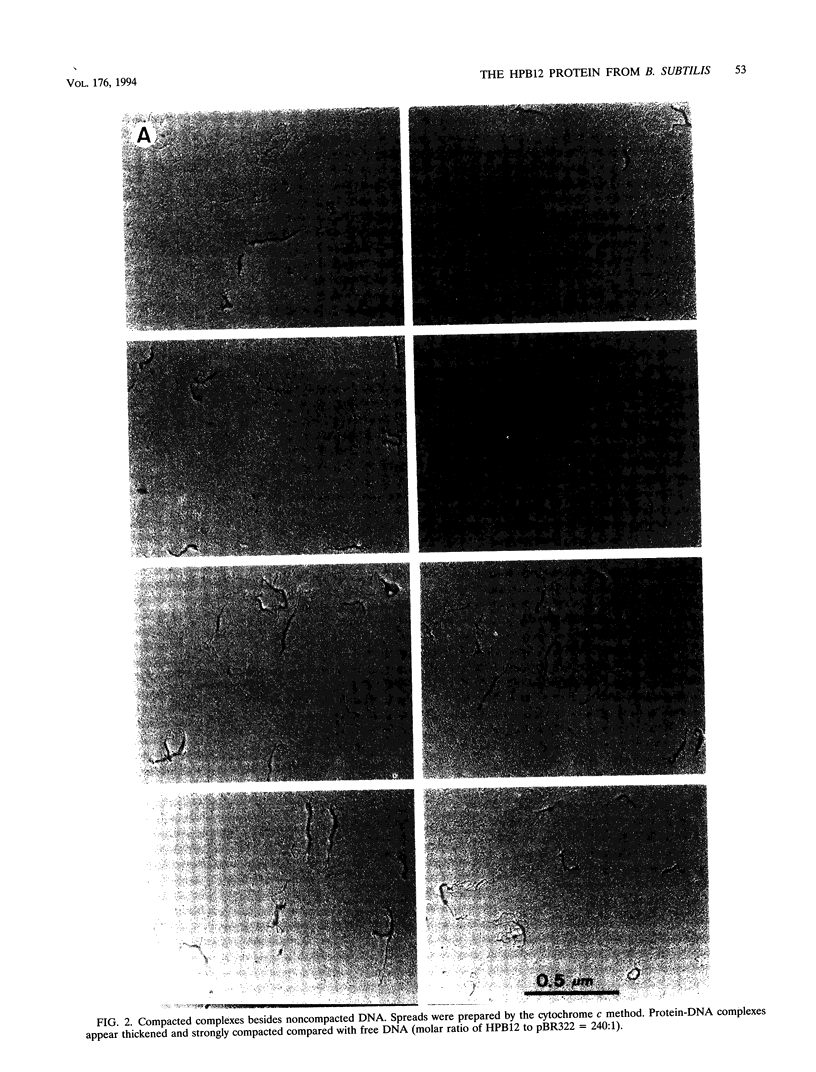
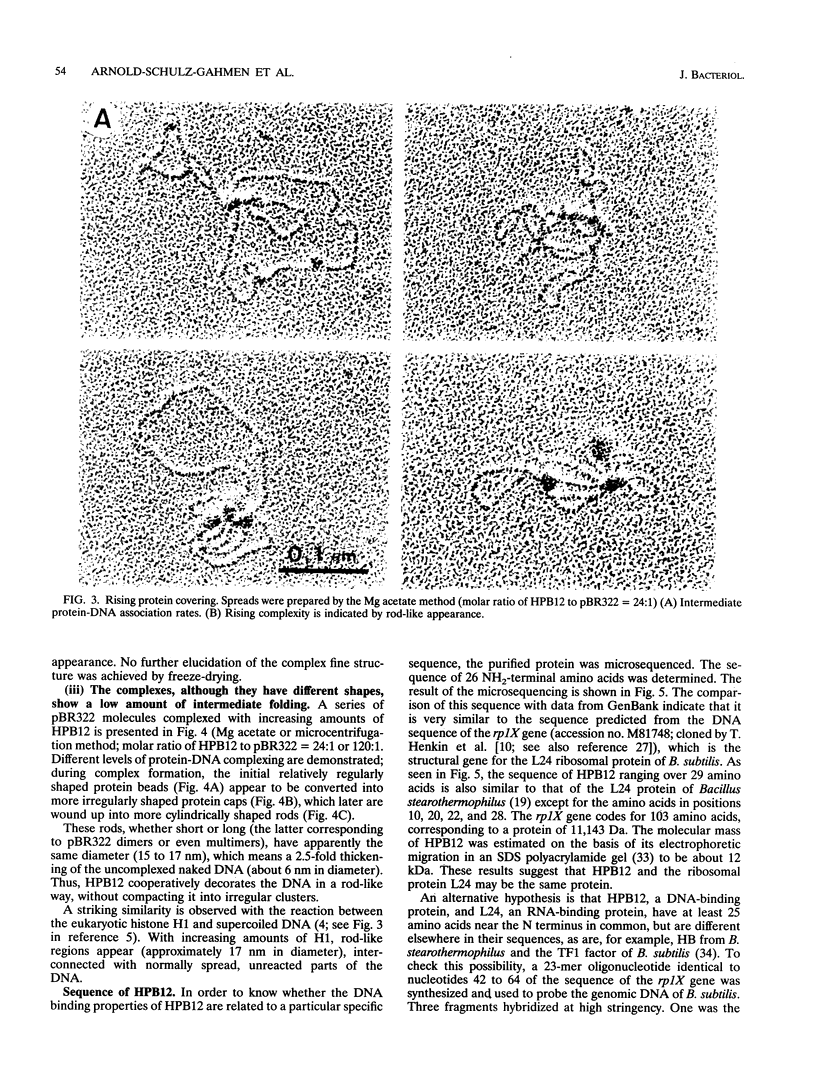
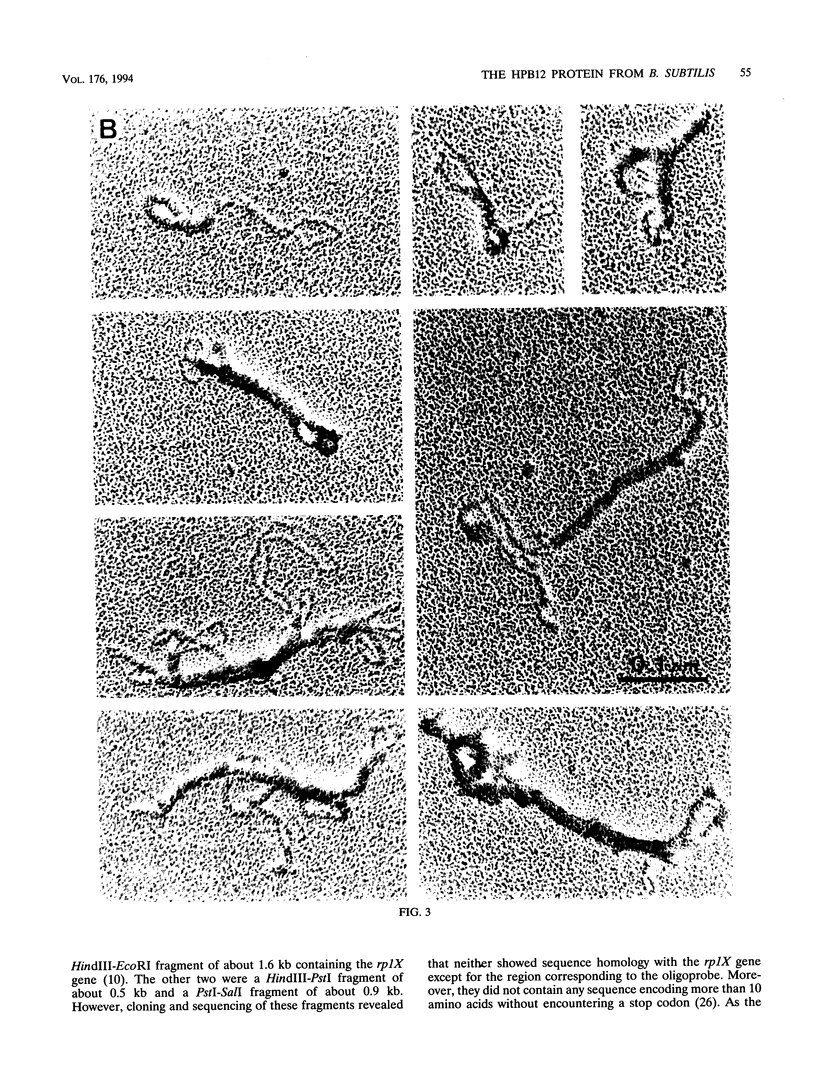
The HPB12 protein from the nucleoid of Bacillus subtilis was previously described, and its DNA binding properties have been reported previously (V. Salti, F. Le Hégarat, and L. Hirschbein, Biochim. Biophys. Acta 1009:161-167, 1989). The DNA-HPB12 complexes were examined by electron microscopy. They appeared as short, slightly curved rods whereas naked DNA showed no compaction. Since only a small number of complexes with an intermediate degree of folding were observed, it appears that the nucleoid-associated protein HPB12 binds cooperatively to DNA, confirming Salti et al. (V. Salti, F. Le Hégarat, and L. Hirschbein, Biochim. Biophys. Acta 1009:161-167, 1989), and gives rise to a tightly compacted DNA-protein complex. N-terminal sequencing of purified HPB12 showed that all but one of the first 26 amino acids were identical to those of the L24 ribosomal protein.
Full text
PDF

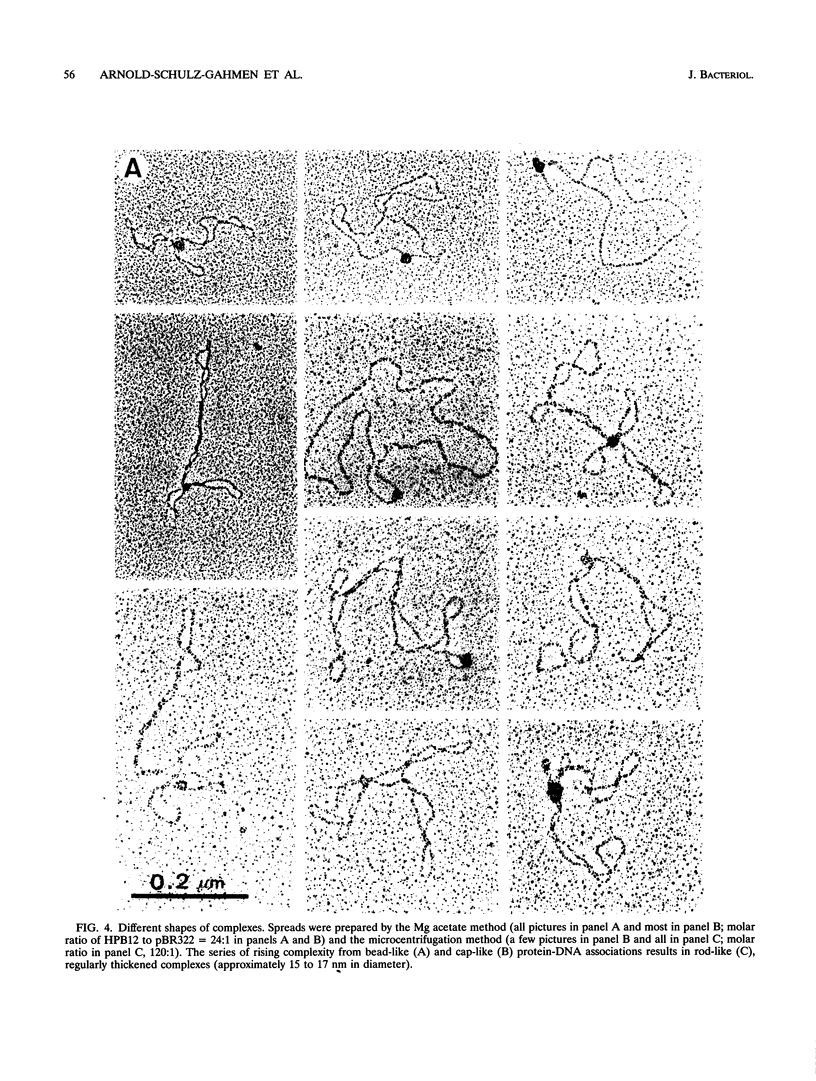
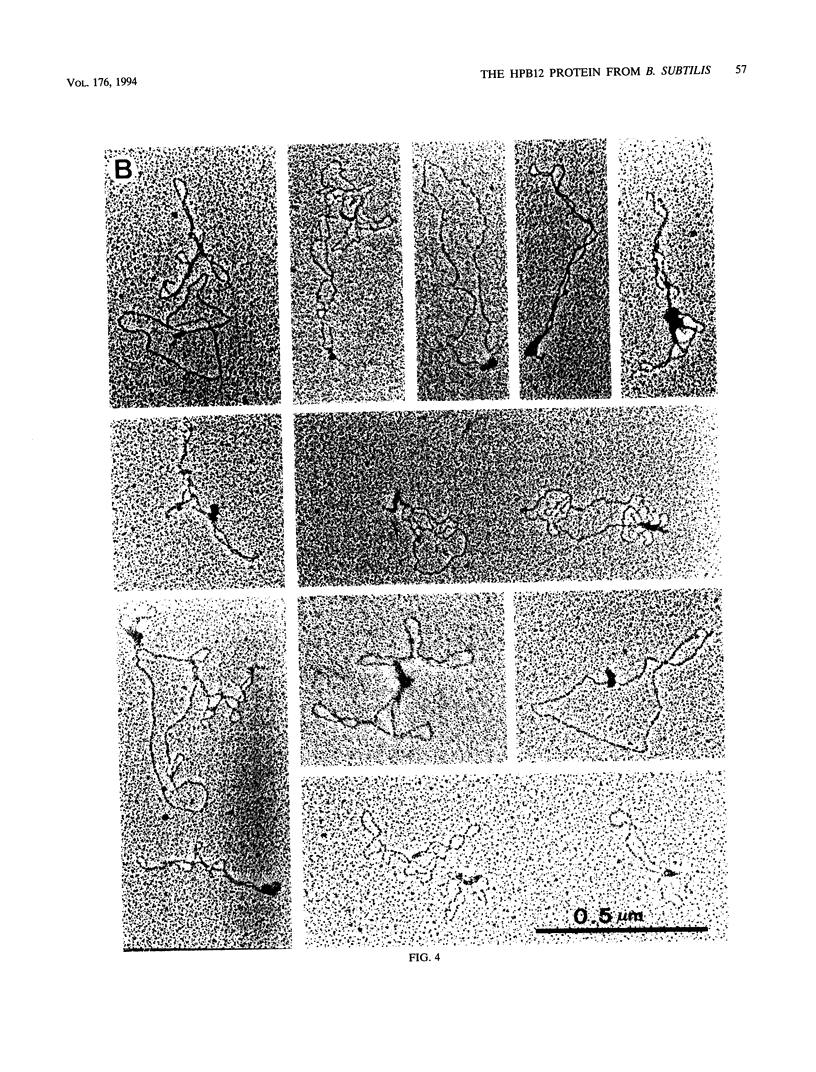
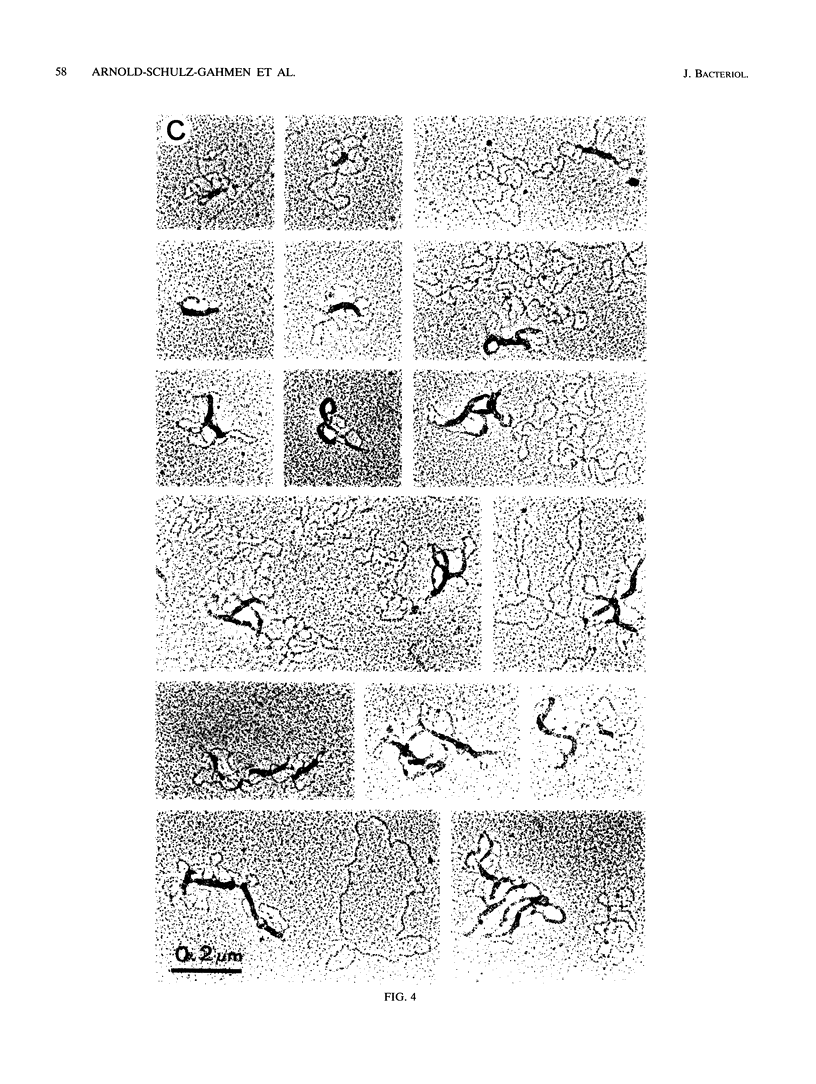


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruckner R. C., Cox M. M. The histone-like H protein of Escherichia coli is ribosomal protein S3. Nucleic Acids Res. 1989 Apr 25;17(8):3145–3161. doi: 10.1093/nar/17.8.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardin W., Losa R., Koller T. Formation and characterization of soluble complexes of histone H1 with supercoiled DNA. J Mol Biol. 1986 Jun 5;189(3):503–517. doi: 10.1016/0022-2836(86)90320-7. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisser M., Tischendorf G. W., Stöffler G. Comparative immunological and electrophoretic studies on ribosomal proteins of bacillaceae. Mol Gen Genet. 1973 Dec 20;127(2):129–145. doi: 10.1007/BF00333661. [DOI] [PubMed] [Google Scholar]
- Grabowski D. T., Deutsch W. A., Derda D., Kelley M. R. Drosophila AP3, a presumptive DNA repair protein, is homologous to human ribosomal associated protein P0. Nucleic Acids Res. 1991 Aug 11;19(15):4297–4297. doi: 10.1093/nar/19.15.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen N., Le Hegaret F., Fleury A. M., Hirschbein L. Folded chromosomes of vegetative Bacillus subtilis: composition and properties. Nucleic Acids Res. 1978 Feb;5(2):475–489. doi: 10.1093/nar/5.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M., Moon S. H., Mattheakis L. C., Nomura M. Cloning and analysis of the spc ribosomal protein operon of Bacillus subtilis: comparison with the spc operon of Escherichia coli. Nucleic Acids Res. 1989 Sep 25;17(18):7469–7486. doi: 10.1093/nar/17.18.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye H., Pollack Y., Petre J. Physical and functional homology between ribosomal protein S1 and interference factor i. Eur J Biochem. 1974 Jun 1;45(1):109–117. doi: 10.1111/j.1432-1033.1974.tb03535.x. [DOI] [PubMed] [Google Scholar]
- Jay G., Kaempfer R. Host interference with viral gene expression: mode of action of bacterial factor i. J Mol Biol. 1974 Jan 15;82(2):193–212. doi: 10.1016/0022-2836(74)90341-6. [DOI] [PubMed] [Google Scholar]
- Kellenberger E., Arnold-Schulz-Gahmen B. Chromatins of low-protein content: special features of their compaction and condensation. FEMS Microbiol Lett. 1992 Dec 15;100(1-3):361–370. doi: 10.1111/j.1574-6968.1992.tb14064.x. [DOI] [PubMed] [Google Scholar]
- Kimura M., Kimura J., Ashman K. The complete primary structure of ribosomal proteins L1, L14, L15, L23, L24 and L29 from Bacillus stearothermophilus. Eur J Biochem. 1985 Aug 1;150(3):491–497. doi: 10.1111/j.1432-1033.1985.tb09049.x. [DOI] [PubMed] [Google Scholar]
- Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9(3):373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- Le Hégarat F., Salti-Montesanto V., Hauck Y., Hirschbein L. Purification and characterization of the HU-like protein HPB9 from the Bacillus subtilis nucleoid. Biochim Biophys Acta. 1993 Feb 20;1172(1-2):101–107. doi: 10.1016/0167-4781(93)90275-i. [DOI] [PubMed] [Google Scholar]
- Mason S. W., Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991 Aug;5(8):1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- Micka B., Groch N., Heinemann U., Marahiel M. A. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J Bacteriol. 1991 May;173(10):3191–3198. doi: 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. L., Jr, Bakken A. H. Morphological studies of transcription. Acta Endocrinol Suppl (Copenh) 1972;168:155–177. doi: 10.1530/acta.0.071s155. [DOI] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Padas P. M., Wilson K. S., Vorgias C. E. The DNA-binding protein HU from mesophilic and thermophilic bacilli: gene cloning, overproduction and purification. Gene. 1992 Aug 1;117(1):39–44. doi: 10.1016/0378-1119(92)90487-a. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E., Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E. Histone-like proteins and bacterial chromosome structure. J Biol Chem. 1988 Sep 15;263(26):12793–12796. [PubMed] [Google Scholar]
- Röhl R., Nierhaus K. H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salti V., Le Hégarat F., Fontaine Y., Hirschbein L. Purification and properties of the DNA-binding protein HPB12 from Bacillus subtilis nucleoid. Biochim Biophys Acta. 1989 Nov 2;1009(2):161–167. doi: 10.1016/0167-4781(89)90096-1. [DOI] [PubMed] [Google Scholar]
- Salti V., Le Hégarat F., Hirschbein L. Isolation and characterization of small heat-stable acid-soluble DNA-binding proteins from Bacillus subtilis nucleoids. J Gen Microbiol. 1985 Mar;131(3):581–590. doi: 10.1099/00221287-131-3-581. [DOI] [PubMed] [Google Scholar]
- Sayre M. H., Geiduschek E. P. Construction and properties of a temperature-sensitive mutation in the gene for the bacteriophage SPO1 DNA-binding protein TF1. J Bacteriol. 1990 Aug;172(8):4672–4681. doi: 10.1128/jb.172.8.4672-4681.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo H., Furubayashi A., Shimizu M., Miyake M., Imamoto F. Preferential binding of E.coli histone-like protein HU alpha to negatively supercoiled DNA. Nucleic Acids Res. 1992 Apr 11;20(7):1553–1558. doi: 10.1093/nar/20.7.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayana T., Subramanian A. R. Specific association of two homologous DNA-binding proteins to the native 30-S ribosomal subunits of Escherichia coli. Biochim Biophys Acta. 1978 Sep 27;520(2):342–357. doi: 10.1016/0005-2787(78)90232-0. [DOI] [PubMed] [Google Scholar]
- Yancey J. E., Matson S. W. The DNA unwinding reaction catalyzed by Rep protein is facilitated by an RHSP-DNA interaction. Nucleic Acids Res. 1991 Jul 25;19(14):3943–3951. doi: 10.1093/nar/19.14.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]