Abstract
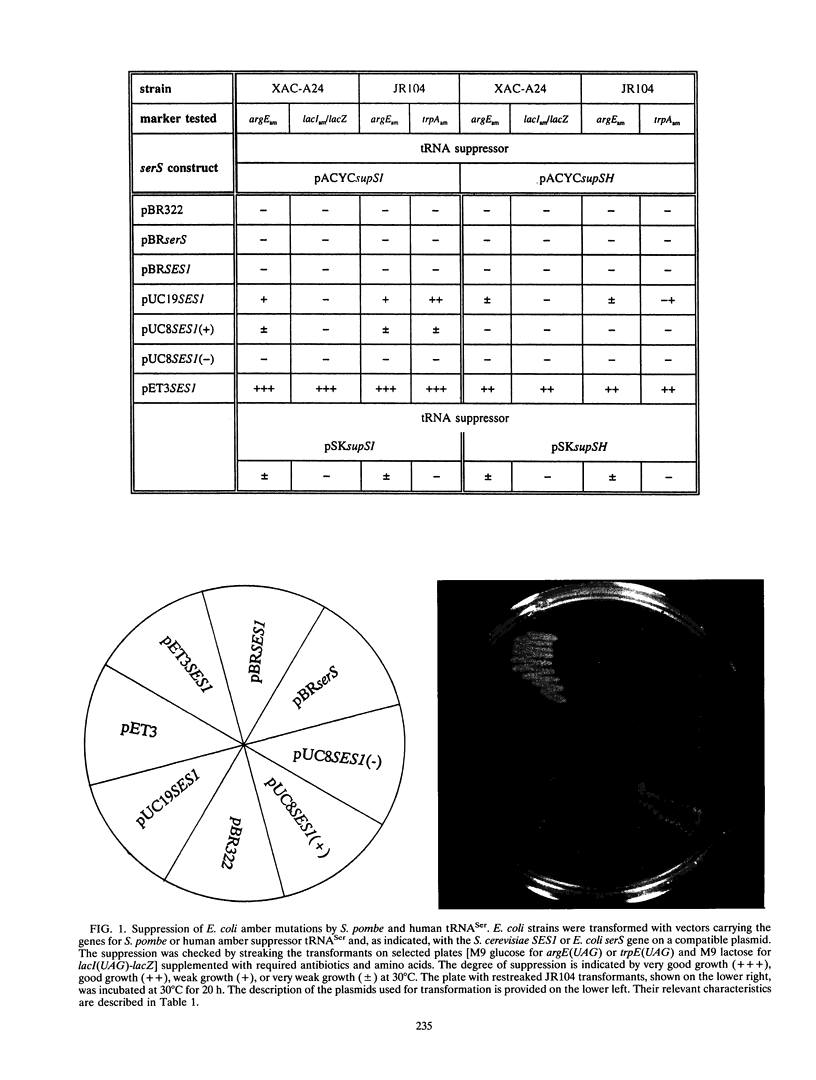
In order to gain insight into the conservation of determinants for tRNA identity between organisms, Schizosaccharomyces pombe and human amber suppressor serine tRNA genes have been examined for functional expression in Escherichia coli. The primary transcripts, which originated from E. coli plasmid promoters, were processed into mature tRNAs, but they were poorly aminoacylated in E. coli and thus were nonfunctional as suppressors in vivo. However, coexpression of cloned Saccharomyces cerevisiae seryl-tRNA synthetase led to efficient suppression in E. coli. This shows that some, but not all, determinants specifying the tRNASer identity are conserved in evolution.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Capone J. P., Sharp P. A., RajBhandary U. L. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 1985 Jan;4(1):213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOCTOR B. P., MUDD J. A. SPECIES SPECIFICITY OF AMINO ACID ACCEPTOR RIBONUCLEIC ACID AND AMINOACYL SOLUBLE RIBONUCLEIC ACID SYNTHETASES. J Biol Chem. 1963 Nov;238:3677–3681. [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Garcia A., Giegé R., Moras D. The contacts of yeast tRNA(Ser) with seryl-tRNA synthetase studied by footprinting experiments. Eur J Biochem. 1990 Mar 10;188(2):283–290. doi: 10.1111/j.1432-1033.1990.tb15401.x. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Edwards H., Schimmel P. A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol Cell Biol. 1990 Apr;10(4):1633–1641. doi: 10.1128/mcb.10.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards H., Trézéguet V., Schimmel P. An Escherichia coli tyrosine transfer RNA is a leucine-specific transfer RNA in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1153–1156. doi: 10.1073/pnas.88.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin S., Lin T. Y., Folk W. R. Construction and expression of nonsense suppressor tRNAs which function in plant cells. Plant J. 1992 Jul;2(4):583–588. doi: 10.1046/j.1365-313x.1992.t01-27-00999.x. [DOI] [PubMed] [Google Scholar]
- Hayase Y., Jahn M., Rogers M. J., Sylvers L. A., Koizumi M., Inoue H., Ohtsuka E., Söll D. Recognition of bases in Escherichia coli tRNA(Gln) by glutaminyl-tRNA synthetase: a complete identity set. EMBO J. 1992 Nov;11(11):4159–4165. doi: 10.1002/j.1460-2075.1992.tb05509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. Conversion of aminoacylation specificity from tRNA(Tyr) to tRNA(Ser) in vitro. Nucleic Acids Res. 1990 Dec 11;18(23):6815–6819. doi: 10.1093/nar/18.23.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Kan Y. W. In vivo aminoacylation of human and Xenopus suppressor tRNAs constructed by site-specific mutagenesis. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2185–2188. doi: 10.1073/pnas.84.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989 Aug 22;28(17):6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- Hudziak R. M., Laski F. A., RajBhandary U. L., Sharp P. A., Capecchi M. R. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell. 1982 Nov;31(1):137–146. doi: 10.1016/0092-8674(82)90413-5. [DOI] [PubMed] [Google Scholar]
- Härtlein M., Madern D., Leberman R. Cloning and characterization of the gene for Escherichia coli seryl-tRNA synthetase. Nucleic Acids Res. 1987 Feb 11;15(3):1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. B. Reaction of aminoacyl-tRNA synthetases with heterologous tRNA's. Prog Nucleic Acid Res Mol Biol. 1971;11:461–488. doi: 10.1016/s0079-6603(08)60335-9. [DOI] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Krupp G., Thurianx P., Willis I., Gamulin V., Söll D. First identification of an amber nonsense mutation in Schizosaccharomyces pombe: major differences in the efficiency of homologous versus heterologous yeast suppressor tRNA genes. Mol Gen Genet. 1985;201(1):82–87. doi: 10.1007/BF00397990. [DOI] [PubMed] [Google Scholar]
- Laski F. A., Belagaje R., RajBhandary U. L., Sharp P. A. An amber suppressor tRNA gene derived by site-specific mutagenesis: cloning and function in mammalian cells. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5813–5817. doi: 10.1073/pnas.79.19.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., RajBhandary U. L. Mutants of Escherichia coli initiator tRNA that suppress amber codons in Saccharomyces cerevisiae and are aminoacylated with tyrosine by yeast extracts. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11378–11382. doi: 10.1073/pnas.88.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Murgola E. J. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- Nicholas H. B., Jr, McClain W. H. An algorithm for discriminating sequences and its application to yeast transfer RNA. Comput Appl Biosci. 1987 Sep;3(3):177–181. doi: 10.1093/bioinformatics/3.3.177. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ollick T., Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., Hall B. D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981 Jun 11;291(5815):464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Racher K. I., Kalmar G. B., Borgford T. J. Expression and characterization of a recombinant yeast isoleucyl-tRNA synthetase. J Biol Chem. 1991 Sep 15;266(26):17158–17164. [PubMed] [Google Scholar]
- Roe B., Sirover M., Dudock B. Kinetics of homologous and heterologous aminoacylation with yeast phenylalanyl transfer ribonucleic acid synthetase. Biochemistry. 1973 Oct 9;12(21):4146–4154. doi: 10.1021/bi00745a018. [DOI] [PubMed] [Google Scholar]
- Rogers M. J., Adachi T., Inokuchi H., Söll D. Switching tRNA(Gln) identity from glutamine to tryptophan. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3463–3467. doi: 10.1073/pnas.89.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6627–6631. doi: 10.1073/pnas.85.18.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J. J., Schold M., Larson G. P., Wallace R. B. Functional expression of a yeast ochre suppressor tRNA gene in Escherichia coli. Gene. 1982 Dec;20(3):423–432. doi: 10.1016/0378-1119(82)90211-6. [DOI] [PubMed] [Google Scholar]
- Roy K. L., Söll D. Purification of five serine transfer ribonucleic acid species from Escherichia coli and their acylation by homologous and heterologous seryl transfer ribonucleic acid synthetases. J Biol Chem. 1970 Mar 25;245(6):1394–1400. [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman J. M., Rogers M. J., Söll D. Competition of aminoacyl-tRNA synthetases for tRNA ensures the accuracy of aminoacylation. Nucleic Acids Res. 1992 Jun 11;20(11):2847–2852. doi: 10.1093/nar/20.11.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundharadas G., Katze J. R., Söll D., Konigsberg W., Lengyel P. On the recognition of serine transfer RNA's specific for unrelated codons by the same seryl-transfer RNA synthetase. Proc Natl Acad Sci U S A. 1968 Oct;61(2):693–700. doi: 10.1073/pnas.61.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvers L. A., Rogers K. C., Shimizu M., Ohtsuka E., Söll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993 Apr 20;32(15):3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991 Dec 25;266(36):24712–24718. [PubMed] [Google Scholar]
- Weygand-Durasevic I., Johnson-Burke D., Söll D. Cloning and characterization of the gene coding for cytoplasmic seryl-tRNA synthetase from Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Mar 11;15(5):1887–1904. doi: 10.1093/nar/15.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weygand-Durasević I., Ban N., Jahn D., Söll D. Yeast seryl-tRNA synthetase expressed in Escherichia coli recognizes bacterial serine-specific tRNAs in vivo. Eur J Biochem. 1993 Jun 15;214(3):869–877. doi: 10.1111/j.1432-1033.1993.tb17990.x. [DOI] [PubMed] [Google Scholar]