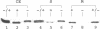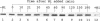Abstract
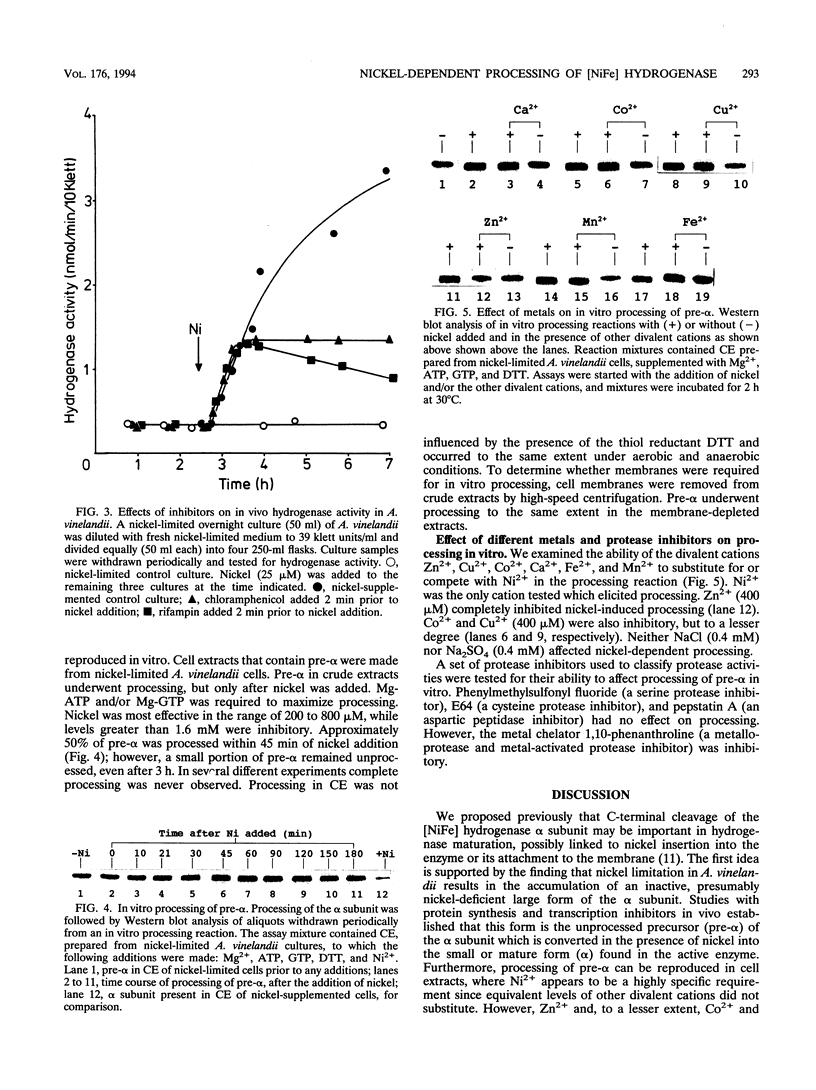
H2 oxidation in Azotobacter vinelandii is catalyzed by a membrane-bound, alpha beta dimeric [NiFe] hydrogenase. Maturation of the enzyme involves cleavage of a putative N-terminal signal sequence in the beta subunit and removal of 15 amino acids from the C terminus of the alpha subunit. Cells limited for nickel exhibited low hydrogenase activities and contained an apparently large form of the alpha subunit. Addition of nickel to such cells increased hydrogenase activities fivefold over 2 h. The increase in the first hour did not require transcription and translation and correlated with processing of the large form of the alpha subunit (pre-alpha) to the small form (alpha) resembling the alpha subunit from the purified enzyme. In vivo, pre-alpha appeared soluble whereas the majority of alpha was membrane bound. Processing of pre-alpha to alpha was reproduced in vitro in membrane-depleted extracts of nickel-limited cells. Processing specifically required the addition of Ni2+, whereas Co2+, Cu2+, Ca2+, Fe2+, Mn2+, and Zn2+ were ineffective. However, Zn2+, Co2+, and Cu2+ inhibited nickel-dependent processing. Mg-ATP and Mg-GTP stimulated processing, whereas anaerobic conditions and/or the addition of dithiothreitol and sodium dithionite was unnecessary. Processing was not inhibited by the protease inhibitors phenylmethylsulfonyl fluoride, E64, and pepstatin.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen J. C., Mortenson L. E. Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim Biophys Acta. 1992 Jun 15;1131(2):199–202. doi: 10.1016/0167-4781(92)90077-d. [DOI] [PubMed] [Google Scholar]
- Chen J. C., Mortenson L. E. Two open reading frames (ORFs) identified near the hydrogenase structural genes in Azotobacter vinelandii, the first ORF may encode for a polypeptide similar to rubredoxins. Biochim Biophys Acta. 1992 May 7;1131(1):122–124. doi: 10.1016/0167-4781(92)90111-c. [DOI] [PubMed] [Google Scholar]
- DE WITT C. W., ROWE J. A. N,O-Diacetylneuraminic acid and N-acetylneuraminic acid in Escherichia coli. Nature. 1959 Aug 1;184(Suppl 6):381–382. doi: 10.1038/184381b0. [DOI] [PubMed] [Google Scholar]
- Diner B. A., Ries D. F., Cohen B. N., Metz J. G. COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J Biol Chem. 1988 Jun 25;263(18):8972–8980. [PubMed] [Google Scholar]
- Friedrich B., Heine E., Finck A., Friedrich C. G. Nickel requirement for active hydrogenase formation in Alcaligenes eutrophus. J Bacteriol. 1981 Mar;145(3):1144–1149. doi: 10.1128/jb.145.3.1144-1149.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G., Schneider K., Friedrich B. Nickel in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1982 Oct;152(1):42–48. doi: 10.1128/jb.152.1.42-48.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Maier R. J. Nickel-dependent reconstitution of hydrogenase apoprotein in Bradyrhizobium japonicum Hupc mutants and direct evidence for a nickel metabolism locus involved in nickel incorporation into the enzyme. Arch Microbiol. 1992;157(6):493–498. doi: 10.1007/BF00276768. [DOI] [PubMed] [Google Scholar]
- Gatenby A. A. Protein folding and chaperonins. Plant Mol Biol. 1992 Jul;19(4):677–687. doi: 10.1007/BF00026793. [DOI] [PubMed] [Google Scholar]
- Gollin D. J., Mortenson L. E., Robson R. L. Carboxyl-terminal processing may be essential for production of active NiFe hydrogenase in Azotobacter vinelandii. FEBS Lett. 1992 Sep 14;309(3):371–375. doi: 10.1016/0014-5793(92)80809-u. [DOI] [PubMed] [Google Scholar]
- Jacobi A., Rossmann R., Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158(6):444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- Kenny B., Taylor S., Holland I. B. Identification of individual amino acids required for secretion within the haemolysin (HlyA) C-terminal targeting region. Mol Microbiol. 1992 Jun;6(11):1477–1489. doi: 10.1111/j.1365-2958.1992.tb00868.x. [DOI] [PubMed] [Google Scholar]
- Klucas R. V., Hanus F. J., Russell S. A., Evans H. J. Nickel: A micronutrient element for hydrogen-dependent growth of Rhizobium japonicum and for expression of urease activity in soybean leaves. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2253–2257. doi: 10.1073/pnas.80.8.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlüke C., Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon A. L., Mortenson L. E., Robson R. L. Nucleotide sequences and genetic analysis of hydrogen oxidation (hox) genes in Azotobacter vinelandii. J Bacteriol. 1992 Jul;174(14):4549–4557. doi: 10.1128/jb.174.14.4549-4557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon A. L., Stults L. W., Robson R. L., Mortenson L. E. Cloning, sequencing and characterization of the [NiFe]hydrogenase-encoding structural genes (hoxK and hoxG) from Azotobacter vinelandii. Gene. 1990 Nov 30;96(1):67–74. doi: 10.1016/0378-1119(90)90342-o. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Peck H. D., Jr, Gall J. L., Przybyla A. E. Cloning and sequencing of the genes encoding the large and small subunits of the periplasmic (NiFeSe) hydrogenase of Desulfovibrio baculatus. J Bacteriol. 1987 Dec;169(12):5401–5407. doi: 10.1128/jb.169.12.5401-5407.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Der Vartanian M., Patil D., Peck H. D., Jr, Menon A. L., Robson R. L., Przybyla A. E. Carboxy-terminal processing of the large subunit of [NiFe] hydrogenases. FEBS Lett. 1993 Sep 27;331(1-2):91–95. doi: 10.1016/0014-5793(93)80303-c. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Wendt J. C., Shanmugam K. T., Przybyla A. E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991 Aug;173(15):4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEWTON J. W., WILSON P. W., BURRIS R. H. Direct demonstration of ammonia as an intermediate in nitrogen fixation by Azotobacter. J Biol Chem. 1953 Sep;204(1):445–451. [PubMed] [Google Scholar]
- Partridge C. D., Yates M. G. Effect of chelating agents on hydrogenase in Azotobacter chroococcum. Evidence that nickel is required for hydrogenase synthesis. Biochem J. 1982 Apr 15;204(1):339–344. doi: 10.1042/bj2040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla A. E., Robbins J., Menon N., Peck H. D., Jr Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992 Feb;8(2):109–135. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993 Mar;57(1):50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., Böhm R., Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992 Jun;6(11):1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Seefeldt L. C., Arp D. J. Purification to homogeneity of Azotobacter vinelandii hydrogenase: a nickel and iron containing alpha beta dimer. Biochimie. 1986 Jan;68(1):25–34. doi: 10.1016/s0300-9084(86)81064-1. [DOI] [PubMed] [Google Scholar]