Abstract
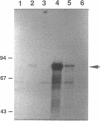
The amino acid sequence of DcrA from Desulfovibrio vulgaris Hildenborough, a strictly anaerobic, sulfate-reducing bacterium, indicated homology with the methyl-accepting chemotaxis proteins from enteric bacteria (A. Dolla, R. Fu, M. J. Brumlik, and G. Voordouw, J. Bacteriol. 174:1726-1733, 1992). The homology is restricted to the cytoplasmic C-terminal signaling domain. The periplasmic N-terminal sensor domain was found to contain a unique sequence, CHHCH, corresponding to a consensus c-type heme binding site. A pretreated, DcrA-specific polyclonal antiserum, generated against DcrA protein overproduced in Escherichia coli, was used for immunoprecipitation of 35S-labeled DcrA from D. vulgaris and Desulfovibrio desulfuricans G200(pJRFR2), a transconjugant that overexpresses functional DcrA. Labeling of the latter with the heme precursor 5-amino-[4-14C]levulinic acid, followed by immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and fluorography, confirmed the presence of c-type heme, while labeling with L-[methyl-3H]methionine in the absence of protein synthesis confirmed that DcrA is a methyl-accepting protein. The base liability of the incorporated radioactivity indicated methyl ester formation like that occurring in the methyl-accepting chemotaxis proteins of enteric bacteria. L-[methyl-3H]methionine labeling of D. desulfuricans G200(pJRFR2) under different conditions indicated that methyl labeling of DcrA decreased upon addition of oxygen and increased upon subsequent addition of the reducing agent dithionite. These results indicate that DcrA may serve as a sensor of oxygen concentration and/or redox potential.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P., Parkinson J. S. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988 Dec 2;55(5):817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Simon M. I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990 Dec 21;63(6):1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Bertrand P., More C., Leroy G., Bonicel J., Haladjian J., Chottard G., Pollock W. B., Voordouw G. Biochemical and spectroscopic characterization of the high molecular weight cytochrome c from Desulfovibrio vulgaris Hildenborough expressed in Desulfovibrio desulfuricans G200. Biochemistry. 1992 Mar 31;31(12):3281–3288. doi: 10.1021/bi00127a033. [DOI] [PubMed] [Google Scholar]
- Chelsky D., Gutterson N. I., Koshland D. E., Jr A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal Biochem. 1984 Aug 15;141(1):143–148. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51(2-3):275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- DeFranco A. L., Koshland D. E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci U S A. 1980 May;77(5):2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckers H. M., Voordouw G. Identification of a large family of genes for putative chemoreceptor proteins in an ordered library of the Desulfovibrio vulgaris Hildenborough genome. J Bacteriol. 1994 Jan;176(2):351–358. doi: 10.1128/jb.176.2.351-358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolla A., Fu R., Brumlik M. J., Voordouw G. Nucleotide sequence of dcrA, a Desulfovibrio vulgaris Hildenborough chemoreceptor gene, and its expression in Escherichia coli. J Bacteriol. 1992 Mar;174(6):1726–1733. doi: 10.1128/jb.174.6.1726-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Finzel B. C., Weber P. C., Hardman K. D., Salemme F. R. Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution. J Mol Biol. 1985 Dec 5;186(3):627–643. doi: 10.1016/0022-2836(85)90135-4. [DOI] [PubMed] [Google Scholar]
- Hamilton W. A. Sulphate-reducing bacteria and anaerobic corrosion. Annu Rev Microbiol. 1985;39:195–217. doi: 10.1146/annurev.mi.39.100185.001211. [DOI] [PubMed] [Google Scholar]
- Harwood C. S. A methyl-accepting protein is involved in benzoate taxis in Pseudomonas putida. J Bacteriol. 1989 Sep;171(9):4603–4608. doi: 10.1128/jb.171.9.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Pollock W. B., Loutfi M., Bruschi M., Rapp-Giles B. J., Wall J. D., Voordouw G. Cloning, sequencing, and expression of the gene encoding the high-molecular-weight cytochrome c from Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1991 Jan;173(1):220–228. doi: 10.1128/jb.173.1.220-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M., Pollock W. B., Reij M. W., Keon R. G., Fu R., Voordouw G. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J Bacteriol. 1993 Aug;175(15):4699–4711. doi: 10.1128/jb.175.15.4699-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi J., Tribhuwan R. C., Berg S. T., Taylor B. L. Signal transduction in chemotaxis to oxygen in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988 Dec;170(12):5507–5511. doi: 10.1128/jb.170.12.5507-5511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Lukat G. S., Stock A. M. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- Taylor B. L. Role of proton motive force in sensory transduction in bacteria. Annu Rev Microbiol. 1983;37:551–573. doi: 10.1146/annurev.mi.37.100183.003003. [DOI] [PubMed] [Google Scholar]
- Unaogu I. C., Gugnani H. C., Lacey J. Occurrence of thermophilic actinomycetes in natural substrates in Nigeria. Antonie Van Leeuwenhoek. 1994;65(1):1–5. doi: 10.1007/BF00878272. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Brenner S. Cloning and sequencing of the gene encoding cytochrome c3 from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem. 1986 Sep 1;159(2):347–351. doi: 10.1111/j.1432-1033.1986.tb09874.x. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Niviere V., Ferris F. G., Fedorak P. M., Westlake D. W. Distribution of Hydrogenase Genes in Desulfovibrio spp. and Their Use in Identification of Species from the Oil Field Environment. Appl Environ Microbiol. 1990 Dec;56(12):3748–3754. doi: 10.1128/aem.56.12.3748-3754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Pollock W. B., Bruschi M., Guerlesquin F., Rapp-Giles B. J., Wall J. D. Functional expression of Desulfovibrio vulgaris Hildenborough cytochrome c3 in Desulfovibrio desulfuricans G200 after conjugational gene transfer from Escherichia coli. J Bacteriol. 1990 Oct;172(10):6122–6126. doi: 10.1128/jb.172.10.6122-6126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Strang J. D., Wilson F. R. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol. 1989 Jul;171(7):3881–3889. doi: 10.1128/jb.171.7.3881-3889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Bartsch R. G., Cusanovich M. A., Hamlin R. C., Howard A., Jordan S. R., Kamen M. D., Meyer T. E., Weatherford D. W., Nguyen huu Xuong Structure of cytochrome c': a dimeric, high-spin haem protein. Nature. 1980 Jul 17;286(5770):302–304. doi: 10.1038/286302a0. [DOI] [PubMed] [Google Scholar]
- Weimer P. J., Van Kavelaar M. J., Michel C. B., Ng T. K. Effect of Phosphate on the Corrosion of Carbon Steel and on the Composition of Corrosion Products in Two-Stage Continuous Cultures of Desulfovibrio desulfuricans. Appl Environ Microbiol. 1988 Feb;54(2):386–396. doi: 10.1128/aem.54.2.386-396.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Macnab R. M., Imae Y. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol. 1990 Jan;172(1):383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen G. J., Bruschi M., Voordouw G. Cloning and sequencing of the gene encoding cytochrome c553 from Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1989 Jun;171(6):3575–3578. doi: 10.1128/jb.171.6.3575-3578.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg W. A., van Dongen W. M., Veeger C. Reduction of the amount of periplasmic hydrogenase in Desulfovibrio vulgaris (Hildenborough) with antisense RNA: direct evidence for an important role of this hydrogenase in lactate metabolism. J Bacteriol. 1991 Jun;173(12):3688–3694. doi: 10.1128/jb.173.12.3688-3694.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wachenfeldt C., Hederstedt L. Bacillus subtilis 13-kilodalton cytochrome c-550 encoded by cccA consists of a membrane-anchor and a heme domain. J Biol Chem. 1990 Aug 15;265(23):13939–13948. [PubMed] [Google Scholar]