Abstract
The srfA operon of Bacillus subtilis functions in the biosynthesis of the lipopeptide antibiotic surfactin. On the basis of nucleotide sequence and genetic analysis, it is believed to encode three enzymes (E1A, E1B, and E2) that catalyze the incorporation of the surfactin substrate amino acids. Insertion, deletion, and amino acid substitution mutations of srfA were analyzed for subunit composition and activity as determined by assays of both amino acid-dependent ATP-PPi exchange and aminoacyl thioester formation. Insertion mutations in srfAA (encoding E1A, the subunit that incorporates Glu, Leu, and D-Leu) eliminated production and activity of all three enzymes. Deletions within srfAA and extending from srfAA to srfAB (encoding E1B, which incorporates Val, Asp, and D-Leu) abolished the activity and production of all three enzymes. Insertions between srfAA and srfAB and within srfAB eliminate the production and activity of E1B and E2. An insertion mutation in srfAC (encoding E2, which incorporates Leu) abolished the activity of E2 only. Mutations of the active serine in the putative 4'-phosphopantetheine-binding motif of the second and third domains of E1A eliminated thioester formation and severely reduced the ATP-PPi exchange activity of the two domains. However, the same mutation in the first domain of E1B had little effect on Val-dependent ATP-PPi exchange activity but abolished thioester formation. These results indicate that the coding assignments of the srfA genes are srfAA (E1A), srfAB (E1B), and srfAC (E2).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cosmina P., Rodriguez F., de Ferra F., Grandi G., Perego M., Venema G., van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993 May;8(5):821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- D'Souza C., Nakano M. M., Corbell N., Zuber P. Amino-acylation site mutations in amino acid-activating domains of surfactin synthetase: effects on surfactin production and competence development in Bacillus subtilis. J Bacteriol. 1993 Jun;175(11):3502–3510. doi: 10.1128/jb.175.11.3502-3510.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuma S., Fujishima Y., Corbell N., D'Souza C., Nakano M. M., Zuber P., Yamane K. Nucleotide sequence of 5' portion of srfA that contains the region required for competence establishment in Bacillus subtilus. Nucleic Acids Res. 1993 Jan 11;21(1):93–97. doi: 10.1093/nar/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinkauf H., Roskoski R., Jr, Lipmann F. Pantetheine-linked peptide intermediates in gramicidin S and tyrocidine biosynthesis. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2069–2072. doi: 10.1073/pnas.68.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krätzschmar J., Krause M., Marahiel M. A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989 Oct;171(10):5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
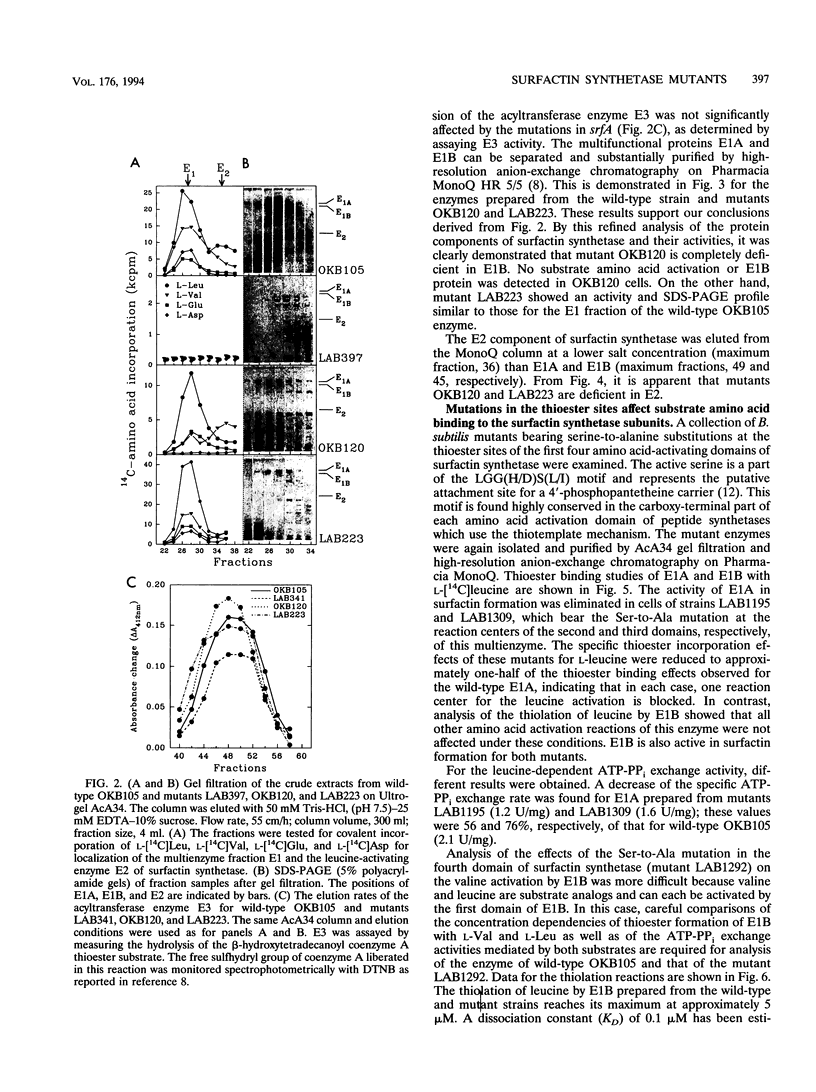
- Menkhaus M., Ullrich C., Kluge B., Vater J., Vollenbroich D., Kamp R. M. Structural and functional organization of the surfactin synthetase multienzyme system. J Biol Chem. 1993 Apr 15;268(11):7678–7684. [PubMed] [Google Scholar]
- Mittenhuber G., Weckermann R., Marahiel M. A. Gene cluster containing the genes for tyrocidine synthetases 1 and 2 from Bacillus brevis: evidence for an operon. J Bacteriol. 1989 Sep;171(9):4881–4887. doi: 10.1128/jb.171.9.4881-4887.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Magnuson R., Myers A., Curry J., Grossman A. D., Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991 Mar;173(5):1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Marahiel M. A., Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988 Dec;170(12):5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumbohm W., Stein T., Ullrich C., Vater J., Krause M., Marahiel M. A., Kruft V., Wittmann-Liebold B. An active serine is involved in covalent substrate amino acid binding at each reaction center of gramicidin S synthetase. J Biol Chem. 1991 Dec 5;266(34):23135–23141. [PubMed] [Google Scholar]
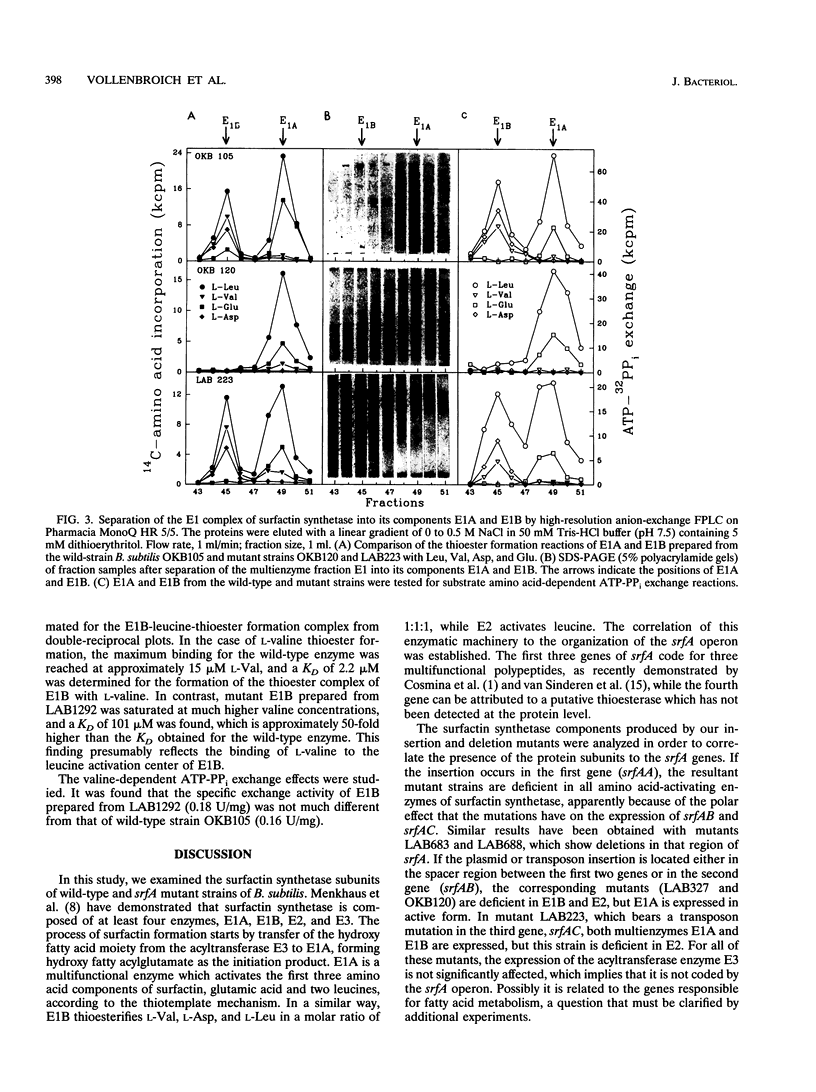
- Turgay K., Krause M., Marahiel M. A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992 Feb;6(4):529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Ullrich C., Kluge B., Palacz Z., Vater J. Cell-free biosynthesis of surfactin, a cyclic lipopeptide produced by Bacillus subtilis. Biochemistry. 1991 Jul 2;30(26):6503–6508. doi: 10.1021/bi00240a022. [DOI] [PubMed] [Google Scholar]
- Vollenbroich D., Kluge B., D'Souza C., Zuber P., Vater J. Analysis of a mutant amino acid-activating domain of surfactin synthetase bearing a serine-to-alanine substitution at the site of carboxylthioester formation. FEBS Lett. 1993 Jul 5;325(3):220–224. doi: 10.1016/0014-5793(93)81077-d. [DOI] [PubMed] [Google Scholar]
- Weckermann R., Fürbass R., Marahiel M. A. Complete nucleotide sequence of the tycA gene coding the tyrocidine synthetase 1 from Bacillus brevis. Nucleic Acids Res. 1988 Dec 23;16(24):11841–11841. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen D., Galli G., Cosmina P., de Ferra F., Withoff S., Venema G., Grandi G. Characterization of the srfA locus of Bacillus subtilis: only the valine-activating domain of srfA is involved in the establishment of genetic competence. Mol Microbiol. 1993 May;8(5):833–841. doi: 10.1111/j.1365-2958.1993.tb01630.x. [DOI] [PubMed] [Google Scholar]