Abstract
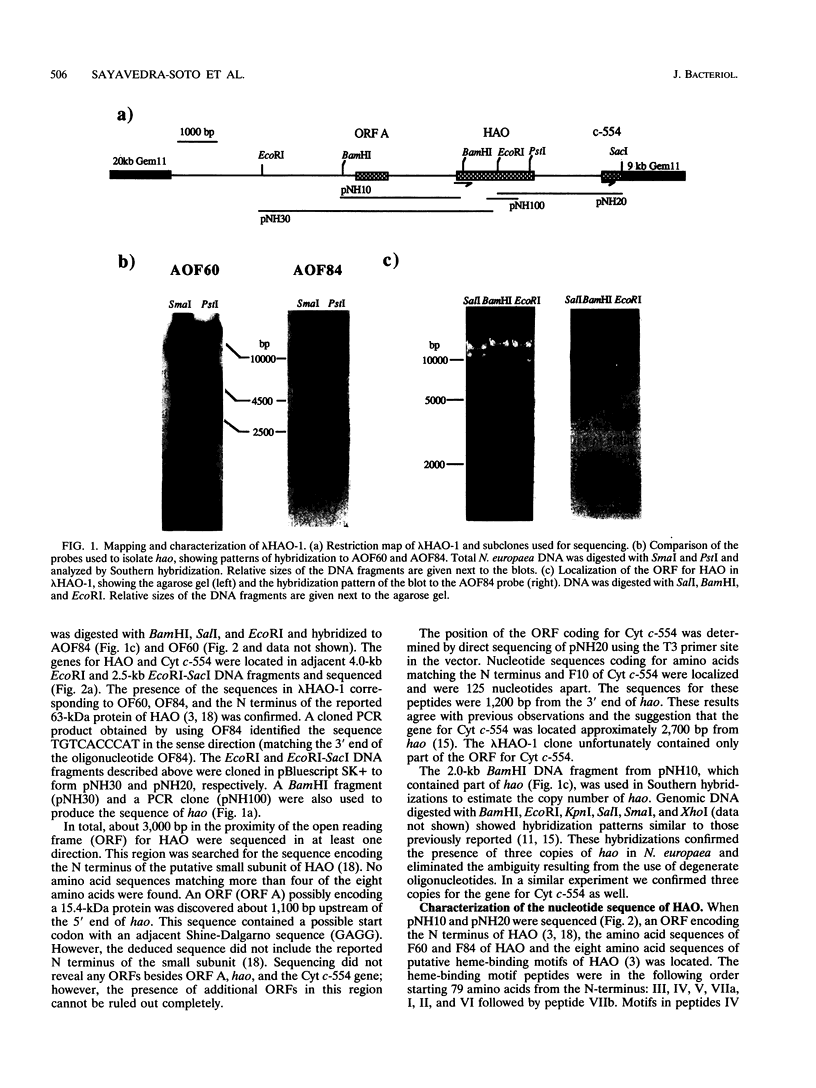
Hydroxylamine oxidoreductase (HAO) catalyzes the oxidation of hydroxylamine to nitrite in Nitrosomonas europaea. The electrons released in the reaction are partitioned to ammonium monooxygenase and to the respiratory chain. The immediate acceptor of electrons from HAO is believed to be cytochrome c-554 (Cyt c-554). We have isolated a genomic DNA fragment containing the structural gene encoding HAO (hao) and a part of the gene for Cyt c-554. The nucleotide sequence of hao was determined, and its transcription was analyzed. The open reading frame (ORF) encodes amino acid sequences matching the purified peptides of HAO. A 64.28-kDa protein is encoded in this ORF, in close agreement with the empirically determined molecular mass of 63 kDa. The N terminus was located 24 amino acids from the start codon, suggesting the presence of a leader sequence. The putative eight heme-binding peptides were localized in this ORF. The gene for Cyt c-554 was located 1,200 bp downstream from the 3' end of hao. An ORF was identified in the upstream region from hao and may encode a protein of unknown function. Data bank searches did not reveal proteins with substantial similarities to HAO, but they did reveal similarities between Cyt c-554 and other c-type cytochromes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Bruschi M., Le Gall J. The structure of cytochrome c'(3) from desulfovibrio gigas (NCIB 9332). FEBS Lett. 1969 Oct 21;5(2):115–117. doi: 10.1016/0014-5793(69)80308-x. [DOI] [PubMed] [Google Scholar]
- Arciero D. M., Balny C., Hooper A. B. Spectroscopic and rapid kinetic studies of reduction of cytochrome c554 by hydroxylamine oxidoreductase from Nitrosomonas europaea. Biochemistry. 1991 Dec 3;30(48):11466–11472. doi: 10.1021/bi00112a014. [DOI] [PubMed] [Google Scholar]
- Arciero D. M., Hooper A. B. Hydroxylamine oxidoreductase from Nitrosomonas europaea is a multimer of an octa-heme subunit. J Biol Chem. 1993 Jul 15;268(20):14645–14654. [PubMed] [Google Scholar]
- Cunliffe V., Koopman P., McLaren A., Trowsdale J. A mouse zinc finger gene which is transiently expressed during spermatogenesis. EMBO J. 1990 Jan;9(1):197–205. doi: 10.1002/j.1460-2075.1990.tb08096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensign S. A., Hyman M. R., Arp D. J. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J Bacteriol. 1993 Apr;175(7):1971–1980. doi: 10.1128/jb.175.7.1971-1980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M., Kress M., Hamer D. A gene that encodes a protein consisting solely of zinc finger domains is preferentially expressed in transformed mouse cells. Mol Cell Biol. 1990 Jan;10(1):418–421. doi: 10.1128/mcb.10.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haser R., Pierrot M., Frey M., Payan F., Astier J. P., Bruschi M., Le Gall J. Structure and sequence of the multihaem cytochrome c3. Nature. 1979 Dec 20;282(5741):806–810. doi: 10.1038/282806a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hooper A. B., Maxwell P. C., Terry K. R. Hydroxylamine oxidoreductase from Nitrosomonas: absorption spectra and content of heme and metal. Biochemistry. 1978 Jul 25;17(15):2984–2989. doi: 10.1021/bi00608a007. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- McTavish H., LaQuier F., Arciero D., Logan M., Mundfrom G., Fuchs J. A., Hooper A. B. Multiple copies of genes coding for electron transport proteins in the bacterium Nitrosomonas europaea. J Bacteriol. 1993 Apr;175(8):2445–2447. doi: 10.1128/jb.175.8.2445-2447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivière V., Wong S. L., Voordouw G. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a beta-lactamase fusion protein. J Gen Microbiol. 1992 Oct;138(10):2173–2183. doi: 10.1099/00221287-138-10-2173. [DOI] [PubMed] [Google Scholar]
- Numata M., Saito T., Yamazaki T., Fukumori Y., Yamanaka T. Cytochrome P-460 of Nitrosomonas europaea: further purification and further characterization. J Biochem. 1990 Dec;108(6):1016–1021. doi: 10.1093/oxfordjournals.jbchem.a123300. [DOI] [PubMed] [Google Scholar]
- Passananti C., Felsani A., Caruso M., Amati P. Mouse genes coding for "zinc-finger"-containing proteins: characterization and expression in differentiated cells. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9417–9421. doi: 10.1073/pnas.86.23.9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M. K. Studies on the hydroxylamine metabolism of Nitrosomonas europaea. II. Molecular properties of the electron-transport particle, hydroxylamine oxidase. Biochemistry. 1968 Jan;7(1):366–372. doi: 10.1021/bi00841a046. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- Shinkai W., Hase T., Yagi T., Matsubara H. Amino acid sequence of cytochrome c3 from Desulfovibrio vulgaris, Miyazaki. J Biochem. 1980 Jun;87(6):1747–1756. doi: 10.1093/oxfordjournals.jbchem.a132919. [DOI] [PubMed] [Google Scholar]
- Terry K. R., Hooper A. B. Hydroxylamine oxidoreductase: a 20-heme, 200 000 molecular weight cytochrome c with unusual denaturation properties which forms a 63 000 molecular weight monomer after heme removal. Biochemistry. 1981 Nov 24;20(24):7026–7032. doi: 10.1021/bi00527a039. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Brenner S. Cloning and sequencing of the gene encoding cytochrome c3 from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem. 1986 Sep 1;159(2):347–351. doi: 10.1111/j.1432-1033.1986.tb09874.x. [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Shinra M. Cytochrome c-552 and cytochrome c-554 derived from Nitrosomonas europaea. Purification, properties, and their function in hydroxylamine oxidation. J Biochem. 1974 Jun;75(6):1265–1273. doi: 10.1093/oxfordjournals.jbchem.a130510. [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Shinra M., Takahashi K., Shibasaka M. Highly purified hydroxylamine oxidoreductase derived from Nitrosomonas europaea. Some physicochemical and enzymatic properties. J Biochem. 1979 Oct;86(4):1101–1108. doi: 10.1093/oxfordjournals.jbchem.a132604. [DOI] [PubMed] [Google Scholar]