Abstract
The Tn611 transposon was inserted into pCG63, a temperature-sensitive plasmid isolated from an Escherichia coli-mycobacterial shuttle vector which contains the pAL5000 and pUC18 replicons. The resulting plasmid, pCG79, was used to generate a large number of insertional mutations in Mycobacterium smegmatis. These are the first mycobacterial insertional mutant libraries to be constructed by transposition directly into a mycobacterium. No highly preferential insertion sites were detected by Southern blot analysis of the chromosomal DNAs isolated from the insertion mutants. Auxotrophic mutants with various phenotypes were isolated at a frequency ranging from 0.1 to 0.4%, suggesting that the libraries are representative. The pCG79 system thus seems to be a useful tool for the study of M. smegmatis genetics and may be applicable to other mycobacteria, such as the M. tuberculosis complex.
Full text
PDF

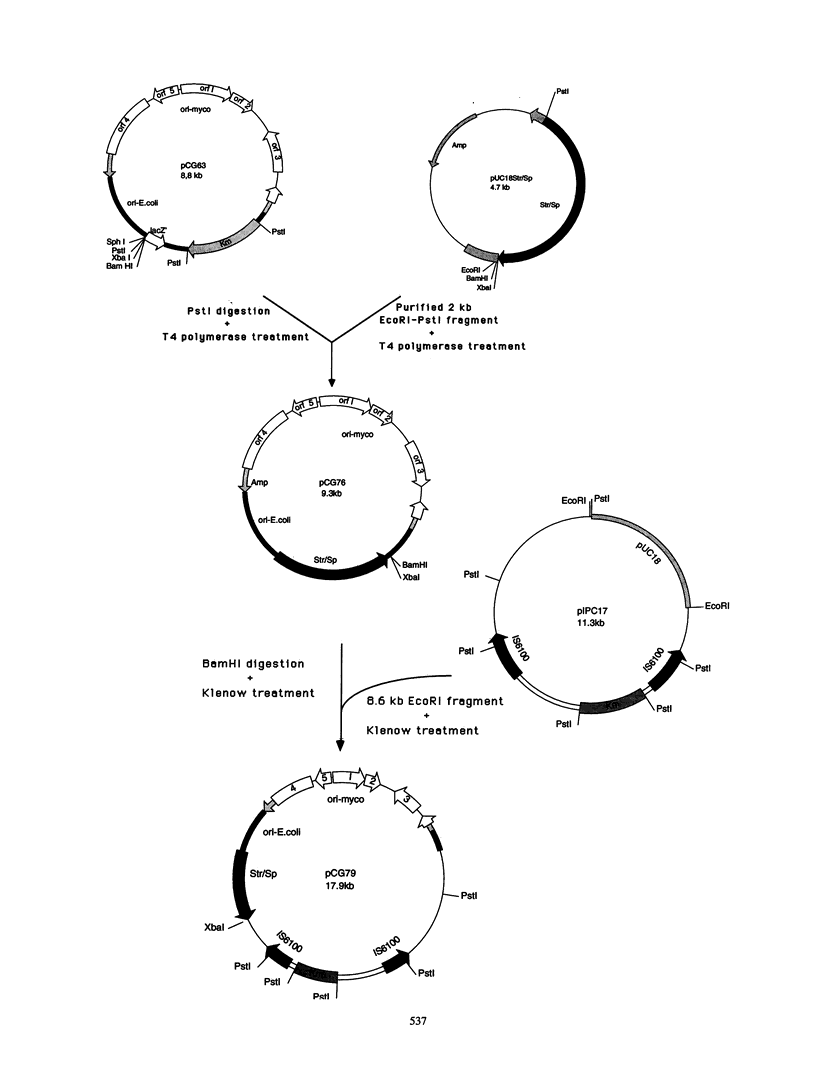
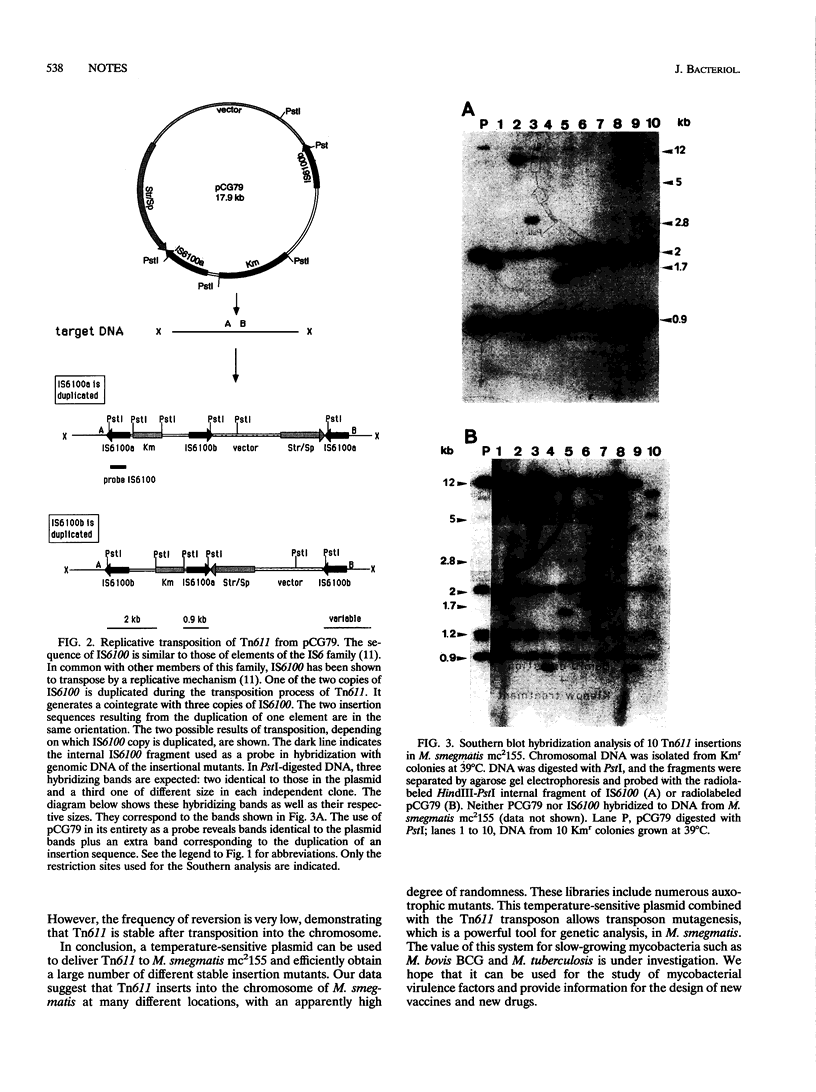

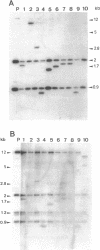
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cirillo J. D., Barletta R. G., Bloom B. R., Jacobs W. R., Jr A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J Bacteriol. 1991 Dec;173(24):7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D. M., Stephens D. M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991 Sep 15;67(1):11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- Fields P. I., Swanson R. V., Haidaris C. G., Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhot C., Gicquel B., Davies J., Martín C. Isolation and analysis of IS6120, a new insertion sequence from Mycobacterium smegmatis. Mol Microbiol. 1992 Jan;6(1):107–113. doi: 10.1111/j.1365-2958.1992.tb00842.x. [DOI] [PubMed] [Google Scholar]
- Guilhot C., Gicquel B., Martín C. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol Lett. 1992 Nov 1;77(1-3):181–186. doi: 10.1016/0378-1097(92)90152-e. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Falkow S. Genetic analysis of bacterial virulence determinants in Bordetella pertussis and the pathogenetic Yersinia. Curr Top Microbiol Immunol. 1985;118:1–11. doi: 10.1007/978-3-642-70586-1_1. [DOI] [PubMed] [Google Scholar]
- Kalpana G. V., Bloom B. R., Jacobs W. R., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Timm J., Rauzier J., Gomez-Lus R., Davies J., Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990 Jun 21;345(6277):739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ranes M. G., Rauzier J., Lagranderie M., Gheorghiu M., Gicquel B. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a "mini" mycobacterium-Escherichia coli shuttle vector. J Bacteriol. 1990 May;172(5):2793–2797. doi: 10.1128/jb.172.5.2793-2797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990 Nov;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]