Abstract
The cytoskeleton of Saccharomyces cerevisiae is essentially invisible using conventional microscopy techniques. A similar problem was solved for the mammalian cell cytoskeleton using resinless section electron microscopy, a technique applied here to yeast. In the resinless image, soluble proteins are no longer cloaked by embedding medium and must be removed by selective detergent extraction. In yeast, this requires breaching the cell wall by digesting with Zymolyase sufficiently to allow detergent extraction of the plasma membrane lipids. Gel electropherograms show that the extracted or “soluble” proteins are distinct from the retained or “structural” proteins that presumably comprise the cytoskeleton. These putative cytoskeleton proteins include the major portions of a 43-kDa protein, which is presumably actin, and of proteins in a band appearing at 55 kDa, as well as numerous less abundant, nonactin proteins. Resinless section electron micrographs show a dense, three-dimensional web of anastomosing, polymorphic filaments bounded by the remnant cell wall. Although the filament network is very heterogenous, there appear to be two principal classes of filament diameters—5 nm and 15–20 nm—which may correspond to actin and intermediate filaments, respectively. A large oval region of lower filament density probably corresponds to the vacuole, and an electron dense spheroidal body, 300–500 nm in diameter, is likely the nucleus. The techniques detailed in this report afford new approaches to the study of yeast cytoarchitecture.
Keywords: cytoarchitecture, detergent extraction, cytoskeletal proteins, Saccharomyces cerevisiae
The yeast cell cytoskeleton has long been the subject of indirect experimentation and considerable speculation (1–7). Studies on yeast cell morphology and cellular dynamics imply an underlying, active organizing structure. However, this structure has never been directly imaged, because conventional microscopy techniques are inadequate. Resinless section electron microscopy, a proven technique for preparing and viewing samples that allows the elucidation of cytoarchitecture in mammalian cells, was modified for the study of the yeast cytoskeleton. This report describes the modifications of the technique required by the very different internal milieu and exterior surface of the yeast cell. The results include, to our knowledge, the first views of the internal yeast cell cytoarchitecture.
Resinless section electron microscopy is a simple, but powerful, extension of conventional electron microscopy techniques. It often is not appreciated that biological material, such as the protein filaments of the cytoskeleton, can form very high-contrast images in the electron microscope by phase interference at the image plane (8). Such images are completely masked by the embedding resin in the conventional ultrathin section. Conventional microscopy images only the heavy-metal atoms adhering to the portion of the sample accessible at the section’s surface. In contrast, the resinless section can image the entire sample in three dimensions and does not require heavy-metal stains. Also, the support of the sample by the embedding medium has proven unnecessary; once chemically fixed, cell structures are strong and possessed of considerable dimensional stability (8, 9).
The first embedment-free micrographs showed a dense complex of soluble proteins that obscured the structural elements of the cytoskeleton (10). To see the cytoskeleton, these soluble proteins—normally cloaked by embedding resin in conventional electron microscopy section (8)—must be removed. This is most easily accomplished by extraction with nonionic detergent in a structure-preserving buffer. The detergent dissolves membrane lipids, thus abolishing the plasma membrane boundary and allowing soluble protein to diffuse away. Adapting this procedure to Saccharomyces cerevisiae requires solving the problem of the impenetrable cell wall and designing a structure-preserving extraction buffer.
MATERIALS AND METHODS
Materials.
Zymolyase 20T was obtained from Seikagaku Kogyo (Tokyo). All other reagents were obtained from Sigma or Mallinckrodt.
Yeast Strain and Cell Growth.
S. cerevisiae of a protease-deficient diploid strain having the genotype bar::LEU2 pep4-3 prb1-1122 prc1-407 was grown in standard yeast extract protein (dextrose 2%) media to mid-log phase by vigorous shaking at 30°C. Enough culture (100–150 ml) was harvested by centrifugation at 3,000 rpm for 5 min to obtain 1 ml of cells.
Partial Enzymatic Digestion of the Cell Wall for Microscopy.
Cells were washed by resuspending them in 6 ml of S buffer (10 mM Pipes, pH 6.5/1.2 M sorbitol/0.5 mM CaCl2) and centrifuging. Unless otherwise noted, all centrifugations were for 5 min at 3,000 rpm at 4°C. Cells were resuspended in 2 ml of S buffer containing 2 μl of 2-mercaptoethanol and incubated at room temperature for 15 min. They were then centrifuged and resuspended in 4 ml of S buffer containing 50 units of Zymolyase (0.5 mg of Zymolyase 100T or 2.5 mg of Zymolyase 20T) and incubated in a shaking bath at 75 rpm, 30°C for 35 min. A more thorough cell wall digestion was used for protein analysis and is described below.
Detergent Extraction of Soluble Proteins.
The digested cells were centrifuged, and the resulting pellet washed twice by resuspending in 12 ml of S buffer and centrifuging again. Cells were then resuspended in 3 ml of yeast cytoskeleton buffer (YCSK) (10 mM Mes, pH 6.0/3 mM MgCl2/1 mM EGTA/0.2 M NaCl/0.8 M sorbitol) containing 0.5% Triton X-100 and a protease inhibitor cocktail [3 μg of leupeptin, 6 μg of pepstatin, 15 μg of aprotinin, and 1.4 mg of AEBSF (Pefabloc, Boehringer Mannheim)] (11), incubated for 5 min at room temperature and centrifuged. The pellet now clearly showed two layers corresponding to distinct populations of cells. The lighter-color upper layer containing only cytoskeletons was aspirated, resuspended in 12 ml of YCSK and centrifuged to wash away remaining soluble proteins.
Preparation of Cells for Resinless Section Electron Microscopy.
The cytoskeletons were resuspended in 3 ml of YCSK containing 2% glutaraldehyde, incubated for 5 min at room temperature, and then centrifuged. The pellet was allowed to incubate further in the glutaraldehyde solution overnight. It subsequently was dehydrated, embedded in modified diethylene glycol distearate (supplied by the EMCorp under the name “Antibed;” the company no longer exists, and an alternative supplier is being sought) and sectioned to a thickness of 90 nm. The sections were placed on Formvar-covered, carbon-coated copper grids, and the embedding material then removed (9). A JEOL 1200 EXII microscope operated at 80 kV was used to view the resinless sections.
Electrophoretic Analysis of Soluble and Cytoskeletal Proteins.
To obtain proteins for electrophoretic analysis, the cell walls were more extensively digested. Cells were harvested as above and washed in 6 ml of S buffer. They were resuspended in 2 ml of S buffer containing 20 μl of β-mercaptoethanol and incubated at room temperature for 15 min. The cells then were centrifuged and resuspended in 4 ml of S buffer containing 400 units of Zymolyase. They were incubated at 75 rpm in a shaking bath at 30°C for 70 min, centrifuged, and then washed twice in 12 ml of S buffer. Cells were resuspended in 3 ml of YCSK containing 0.5% Triton X-100 and the protease inhibitor cocktail, incubated for 5 min at room temperature and then centrifuged in an ultracentrifuge for 90 min at 40,000 rpm. The supernatant had a thin milky-white top layer, probably composed of lipid- and carbohydrate-rich cell fragments, which was removed and discarded. The remaining liquid was made 0.5% SDS. The pellet containing cytoskeletal proteins was resuspended in 1 ml 0.5% SDS solution. The cytoskeletal and soluble proteins were analyzed in a polyacrylamide minigel with Rainbow Marker molecular weight standard (Sigma) and silver-stained.
RESULTS
Detergent Extraction of the Yeast Cell.
The first step was to prepare the yeast cell cytoskeleton by removing the soluble proteins. In mammalian cells, the nonionic detergent Triton X-100, in a suitable buffer, is used to dissolve the plasma membrane lipids and release the soluble proteins. In yeast, the cell wall prevents detergents from reaching the plasma membrane, but enzymatic digestion of the wall with Zymolyase can remove this barrier.
To preserve cytostructure best, the extraction buffer should approximate the normal cell internal ionic milieu. It was found empirically that yeast extraction required modifying the YCSK buffer used for mammalian cells. The pH was lowered to 6.0 from 6.8 to match the internal pH of the S. cerevisiae cell more closely (12). The monovalent salt concentration was raised to 0.2 M, which appeared to give the best preservation of filaments with the least amount of adhering material. To prevent osmotic shock, the solution was made slightly hypertonic to the yeast cell with the addition of 0.8 M sorbitol. The resulting YCSK is described fully in Materials and Methods.
Soluble and Cytoskeletal Proteins.
Detergent extraction should separate cell proteins cleanly into soluble and structure-bound fractions. In mammalian cells, these fractions are distinctly different in composition, and these differences indicate the efficacy of separation. To make a similar measurement in yeast cells, the soluble proteins released into the extraction buffer were compared with the cytoskeletal proteins that sedimented into the pellet. The SDS/PAGE electropherogram in Fig. 1 shows that the soluble and structural fractions are quite different, suggesting an effective separation. Two major bands should be noted at approximately 43 and 55 kDa. The 43-kDa band appears almost entirely in the cytoskeleton fraction and very likely corresponds to yeast actin. The band at approximately 55 kDa, representing one or more of the most abundant cellular proteins, is enriched in the skeleton fraction. Its molecular mass and association with the cytoskeleton suggest that this band includes the yeast tubulins and possibly a yeast analogue of intermediate filament protein.
Figure 1.
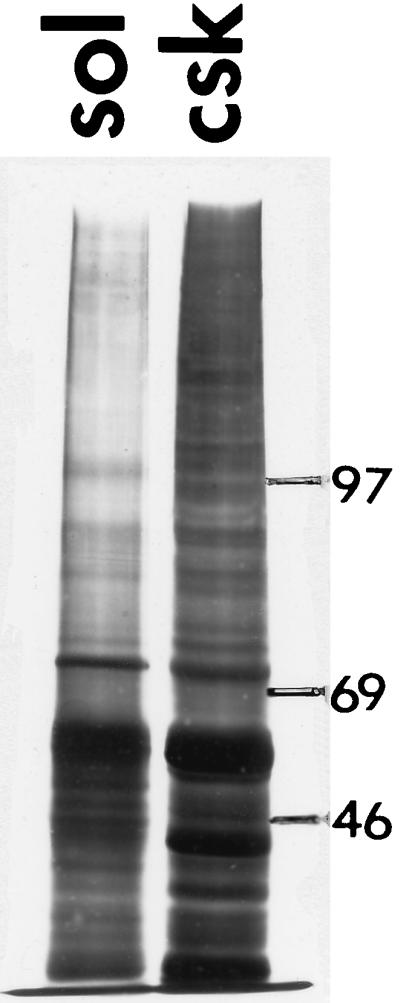
SDS/PAGE electropherogram of structural (Left) and soluble (Right) protein fractions of S. cerevisiae after detergent extraction. Molecular mass markers are on the right.
Electron Microscopy.
The limit digest of cell walls used for protein analysis transforms the cell into a spheroplast, greatly changing cell shape and cytoskeletal architecture. Therefore, to preserve internal architecture better, a partial, nonsphereoplasting, digestion was used in preparing cells for electron microscopy. Not all cell walls were digested sufficiently to allow detergent extraction, but those cells with sufficiently penetrated walls retained enough cell wall fragments to support the cytoskeleton in its original conformation. Examination was restricted to cells that were fully extracted. These formed a lighter-colored layer on the top of the centrifugal pellet and were harvested by gentle aspiration. The cytoskeletons were fixed in glutaraldehyde and resinless sections prepared for microscopy using modified diethylene glycol distearate as a temporary sectioning medium. The yeast cytoskeleton is much denser than that of mammalian cells and requires thinner sections—about 90 nm compared with about 200 nm—to avoid visual confusion.
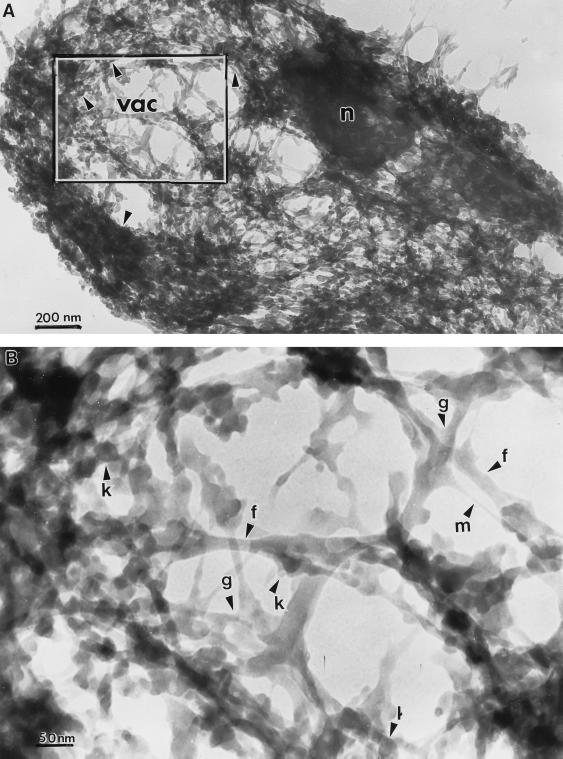
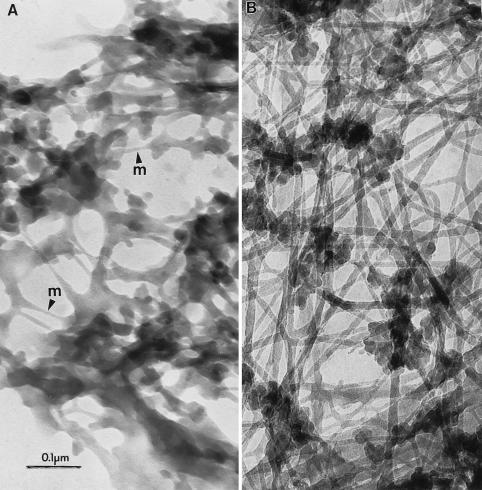
Fig. 2A shows a resinless section micrograph of the S. cerevisiae cytoskeleton at low magnification. It partially resembles the cytoskeleton of mammalian cells in consisting of a complex meshwork of filaments and many other structures that constitute the cytoarchitecture. However, yeast cytoskeletal filaments are much more densely packed than in most mammalian cells and the high magnification micrograph in Fig. 2B shows they are generally much shorter. Fig. 3 compares yeast (A) and mammalian whole-mount (B) cytoskeleton at the same intermediate magnification.
Figure 2.
Resinless section electron micrographs of detergent-extracted S. cerevisiae cells. (A) Whole cytoskeleton showing location of putative nucleus (n), the region likely corresponding to the vacuole (vac), and the area shown at high magnification in B. (Bar = 200 nm.) (B) Detailed view of a portion of the cytoskeleton in A showing fibers resembling the intermediate filaments of mammalian cells (f), thin filaments designated tentatively as microfilaments (m), knob-like bodies (k), and cytoskeletal gussets (g). (Bar = 50 nm.)
Figure 3.
(A) Intermediate-magnification image of a S. cerevisiae cytoskeleton using resinless section electron microscopy. m, thin filaments. (B) Rat aortic smooth muscle cells at similar magnification. (Bar = 100 nm.)
Characteristics of the S. cerevisiae Cytoskeleton.
There are two principal classes of filaments, categorized by diameter, that are visible in the S. cerevisiae cytoskeleton. These can be seen in the high-magnification images of Figs. 2B and 3A. The 5-nm filaments probably are composed of actin and correspond to the 43-kDa protein seen in the cytoskeleton lane in Fig. 1. We tentatively have designated these as “microfilaments.” The 15- to 20-nm filaments may be a yeast analogue of the mammalian intermediate filaments and possibly are composed of proteins found in the 55-kDa protein band. Microtubules, which have a diameter of about 28 nm, were not stabilized during extraction and apparently are absent from these preparations.
Filament junctions in the mammalian cell cytoskeleton (Fig. 3B) show no obvious specialization. In contrast, such junctions in yeast cells often show gussets or brackets. These structures are especially pronounced when the angle between the filaments is acute. Their electron density is the same as that of the filaments themselves. Whether these junctions are present in the cell or are artifacts of extraction remains to be determined. Also, in contrast to many mammalian cytoskeleton filaments, which are relatively smooth and long, the filaments of the S. cerevisiae cytoskeleton are short, polymorphic along their length, and studded with knob-like bodies about 40 nm in diameter. These knob-like structures are often more electron dense than the filaments with which they are associated, suggesting they may contain cytoplasmic RNA.
The cytoskeleton of S. cerevisiae revealed here has some gross morphological features worth noting. It is bounded by the undigested wall fragments. It is markedly inhomogeneous, with regions of varying density throughout the cell. Many sections contain an oval area of much lower filament density, such as that seen in Fig. 2A, that probably corresponds to the cell vacuole. It is significant that these large, low-density areas are never completely empty, but always contain some filaments, suggesting that some class of cytoskeleton fibers extends into the vacuolar space. This finding is consistent with a previous report of cytoskeleton-bound proteins associated with the S. cerevisiae vacuole (6). Often the sections show a roughly spherical region, 300 to 500 nm in diameter, of high electron density (Fig. 2A). The contained material is so electron dense as to appear completely opaque. This region is most probably the nucleus; the dense material, chromatin.
DISCUSSION
This report shows the feasibility of adapting the resinless section technique, developed for mammalian cells, to the study of yeast cytoarchitecture. The procedures are straightforward, require no special skills, and afford high-contrast, three-dimensional images of unequaled resolution. Many informative applications of this technique are apparent. These include immunogold staining to localize antigens bound to the cytoskeleton and observation of cytoskeletal effects of morphology-altering treatments. Through resinless section electron microscopy, morphological studies finally can be added to the tools necessary for understanding the cytoskeleton and cytoarchitecture of S. cerevisiae.
Acknowledgments
We thank Prof. Peter Sorger of the Massachusetts Institute of Technology Biology Department and the members of his staff for their invaluable assistance in obtaining and culturing S. cerevisiae and for giving an indispensable introduction to yeast biology. Special thanks are due Iain Russell, who offered the invaluable suggestion of using partial wall digests for morphological studies. We thank Katherine Wan for excellent technical support and advice, Gabriella Krockmalnic for an introduction to the arcane arts of electron microscopy, and Jeffrey Nickerson for guidance in experimental procedures. This research was made possible by National Institutes of Health Grants CA-67628-30 and AR-42262.
ABBREVIATION
- YCSK
yeast cytoskeleton buffer
References
- 1.Wang T, Bretscher A. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman N L, Lila T, Mintzer K A, Chen Z, Pahk A J, Ren R, Drubin D G, Field J. Mol Cell Biol. 1996;16:548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ursic D, Sedbrook J C, Himmel K L, Culbertson M R. Mol Biol Cell. 1994;5:1065–1080. doi: 10.1091/mbc.5.10.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly S F, Pocklington M J, Pallotta D, Orr D. Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 5.McConnell S J, Yaffe M P. Science. 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- 6.Horazdovsky B F, Emr S D. J Biol Chem. 1993;268:4953–4962. [PubMed] [Google Scholar]
- 7.Hyman A A, Sorger P K. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 8.Penman S. Proc Natl Acad Sci USA. 1995;92:5251–5257. doi: 10.1073/pnas.92.12.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capco D G, Krochmalnic G, Penman S. J Cell Biol. 1984;98:1878–1885. doi: 10.1083/jcb.98.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter K R. J Cell Biol. 1984;99:3s–12s. doi: 10.1083/jcb.99.1.3s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones E W. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 12.Pena A, Ramirez J, Rosas G, Calahorra M. J Bacteriol. 1995;177:1017–1022. doi: 10.1128/jb.177.4.1017-1022.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]