Abstract
Ran is a small GTPase required for nuclear transport in eukaryotic cells [Gorlich, D. & Mattaj, I. W. (1996) Science 271, 1513–1518]. Mutants in Ran also show defects in mRNA processing, cell cycle regulation, and other aspects of nuclear function [Rush, M. G., Drivas, G. & D’Eustachio, P. (1996) BioEssays 18, 103–112; Sazer, S. (1996) Trends Cell Biol. 6, 81–85]. In an effort to understand the role of Ran in these diverse processes, we previously characterized 10 Ran interacting proteins (Rips) from Xenopus egg extracts. In this report, we present further characterization of a complex containing three of these Rips: p340RanBP2, p88, and p18. We have cloned the Xenopus homologue of RanGAP1, and we show here that p88 is a modified form of this protein. In RanGAP assays, the p340RanBP2–p88–p18 complex contains GTPase-activating protein activity, indicating that RanGAP1 is not inactivated by modification. Rather, modification of RanGAP1 appears to be linked to its association with p340RanBP2 because we did not observe unmodified RanGAP1 in p340RanBP2 immunoprecipitates. We have also characterized p18, and we found that it is the Xenopus homologue of Ubc9p, an E2 ubiquitin-conjugating enzyme that is required for cell cycle regulation [Seufert, W., Futcher, B. & Jentsch, S. (1995) Nature (London) 373, 78–81]. Using antibodies directed against Xenopus Ubc9p, we have confirmed that Ubc9p associates with p340RanBP2 in Xenopus extracts. These results suggest Ubc9p’s role in cell cycle regulation may involve either modification of nuclear transport substrates or the nuclear transport machinery.
Ran is a small GTPase of the Ras superfamily that is essential for nuclear transport, mRNA processing, maintenance of structural integrity of nuclei, and cell cycle control (1–3). The best characterized role of Ran is in nuclear protein import, and multiple lines of evidence suggest that GTP hydrolysis by Ran is required to sustain both active protein import and export (1).
Components of the Ran GTPase pathway are also required to regulate the entry into mitosis with respect to the completion of DNA replication. This was first observed in tsBN2 cells, a temperature sensitive BHK cell line that has a mutant allele of Ran’s guanine nucleotide exchange factor, RCC1 (4). At the restrictive temperature, S phase tsBN2 cells enter mitosis and undergo premature chromosome condensation and nuclear envelope breakdown despite incomplete DNA replication (5). In yeast, mutants of Ran, RCC1, and RanBP1, a Ran binding protein, show phenotypes that are consistent with a requirement for these proteins in mitotic regulation (6). Experiments in the Xenopus in vitro system have suggested that Ran’s role in regulating mitosis is distinct from its role in nuclear transport; Xenopus egg extracts mimic cell cycle transitions of the early embryo, allowing an examination of the effects of mutant Ran proteins on the regulation of mitosis (7, 8). In cycling extracts, mutant Ran proteins block the activation of cyclin B/p34cdc2 as a mitotic kinase in the absence of nuclear DNA, indicating that Ran regulates mitosis in a manner that is independent of nuclear transport.
Another protein that has been implicated in regulating the onset of mitosis is Ubc9p, a nuclear E2 ubiquitin-conjugating enzyme (9). E2 ubiquitin-conjugating enzymes mediate the attachment of ubiquitin to a variety of protein substrates (10). In Saccharomyces cerevisiae, repression of Ubc9p synthesis blocks cell cycle progression in late G2 or early M phase, and results in the stabilization of B-type cyclins (9). However, Ubc9p is not able to ubiquinate mitotic cyclins in vitro, and it is unlikely to be the enzyme that directly modifies them for destruction during the cell cycle (11). Mammalian homologues of Ubc9p have been cloned in two-hybrid screens by virtue of their association with the recombination protein Rad51p (12), the CBF3 centromere-binding complex from S. cerevisiae (13), the papillomavirus E1 protein (14), and the adenovirus E1A protein (15).
We previously documented the association of three Ran-interacting proteins in Xenopus extracts as a complex that was independent of the nucleotide binding state of Ran (16). This complex contained p340RanBP2 (the Xenopus homologue of RanBP2/Nup358), p88 (a RanGAP1/Fug1-related protein), and p18 (an unknown protein). Mammalian RanBP2/Nup358 is a large nucleoporin with a leucine-rich domain, four RanBP1-related domains, a region of cyclophilin homology, and eight zinc fingers (17, 18). RanBP2/Nup358 has been implicated as the site of protein import substrate docking at the nuclear pore before translocation (19). RanGAP1/Fug1 is a GTPase-activating protein (GAP) for Ran (20). Mutants in RanGAP1 also show pleiotropic phenotypes, with disruption of mRNA processing, nuclear transport, and nuclear structure (21). The association of p340RanBP2 and p88 was suggestive of a model wherein p88-facilitated Ran-GTP hydrolysis might promote the translocation of docked substrates.
In this report, we discuss the further characterization of the p340RanBP2–p88–p18 complex. We have cloned the Xenopus homologue of RanGAP1 and raised antibodies against it. We have found that p88 is a modified form of this protein, and we demonstrate that it is active as a GAP for Ran. Modification of RanGAP1 appears to be linked to its association with RanBP2 because we did not observe unmodified RanGAP1 in anti-p340RanBP2 immunoprecipitates. We have also purified p18 and subjected it to protein sequence analysis, and we found that it is the Xenopus homologue of Ubc9p. We cloned Xenopus Ubc9p and generated rabbit polyclonal antibodies directed against it. Using these antibodies, we confirmed that Ubc9p associates with p340RanBP2 and p88 in Xenopus extracts. These results suggest that Ubc9p’s role in cell cycle regulation may include the ubiquitination of nuclear proteins, perhaps in a manner that is coupled to nuclear transport, or the ubiquitination of the nuclear transport machinery itself.
MATERIALS AND METHODS
Buffers and Reagents.
Buffer A is 20 mM Tris·HCl, pH 8.0/50 mM NaCl/2.5 mM MgCl2/10% glycerol/0.1% Triton X-100/0.1 mM DTT. Column buffer is 50 mM Tris·HCl, pH 8.0/50 mM NaCl/2 mM MgCl2/0.1 mM DTT. The 1× SDS-sample buffer is 50 mM Tris·HCl, pH 6.8/100 mM DTT/2% SDS/0.1% bromophenol blue/10% glycerol. In all figures, proteins were analyzed on 4–20% gradient SDS/polyacrylamide gels (NOVEX, San Diego). Protein concentrations were determined using a Bio-Rad protein assay kit. Other reagents were obtained from Sigma unless otherwise stated.
Preparation of Xenopus Egg Extracts.
Fractionated interphase egg extracts were prepared according to the methods described by Smythe and Newport (22). Cytoplasmic fractions that contained membrane-free soluble proteins were used for the experiments described and are referred to as Xenopus egg extracts throughout the text.
Protein Sequence Analysis of p18.
p18 was prepared from egg extracts through coimmunoprecipitation using anti-p340RanBP2 antibodies (16). Approximately 2 μg of p18 was transferred to nitrocellulose membranes. The proteins on the membranes were visualized by Ponceau S. Determination of peptide sequences was performed at the protein biochemistry facility of SAIC Frederick (Frederick, MD).
Cloning of Xenopus UBC9.
Conserved Ubc9p amino acid sequences FGFVAVP and DWRPAIT were used to design PCR primers. A 270-bp fragment of UBC9 cDNA was obtained by PCR from Xenopus oocyte cDNA. After DNA sequencing of the PCR fragment using a Sequenase kit (United States Biochemicals), the 5′ and 3′ cDNA ends were amplified using the RACE System (GIBCO/BRL). To produce the full-length Xenopus UBC9 cDNA, reverse-transcribed oocyte RNA was used in a PCR with oligonucleotide 1 (5′-agccatatgtctggcatagccctgagcag-3′) and oligonucleotide 2 (5′-gaggatccttatgatggcgcaaacttctt-3′) for the 5′ and 3′ ends, respectively. The resulting 493-bp fragment was subcloned into pCR-II vector (Invitrogen) and sequenced. This clone will be referred to as pcUBC9-3.
Cloning of Xenopus RanGAP1.
Conserved RanGAP1 protein sequences (LSDNAFG and QNGINH) were used to design PCR primers. A reverse transcription–PCR was used to amplify a fragment of RanGAP1 cDNA from Xenopus oocyte RNA. The amplified DNA fragment was subcloned into a pCR-II vector and radiolabeled using a random primer labeling kit purchased from GIBCO/BRL. The radiolabeled fragment was used to screen a Lambda-ZAP-based Xenopus oocyte cDNA library (the kind gift of D. Patterton and A. Wolffe, Laboratory of Molecular Biology, National Institute of Child Health and Human Development), and four positive single plaques were purified. Plasmid DNA was rescued from each of these clones. Three clones were sequenced and found to contain the same DNA sequence. One clone (GAP1-6-1) contained the entire coding region of RanGAP1 and was sequenced on both strands.
Antibodies.
pcUBC9-3 was double-digested by NdeI–BamHI, and the insert DNA was subcloned into NdeI–BamHI site of the pET28a plasmid vector (Novagen). This construct (pHis6-UBC9) gives a protein with a 20-aa extension at the N terminus of Ubc9p, including a His tag. Expression of tagged Ubc9p was induced in Escherichia coli [strain BL21(DE3) pLysS] under standard conditions (16). His-tagged Ubc9p was purified on Ni+2-agarose (Novagen). To prepare anti-Ubc9p antibodies, the purified His-tagged Ubc9p was subjected to SDS/PAGE (15% gel). The band containing the His-tagged Ubc9p was visualized by Coomassie brilliant blue, excised, and used to elicit antibodies in two rabbits (Research Genetics, Huntsville, AL).
The first 360 aa of Xenopus RanGAP1 were amplified by PCR using oligonucleotide 3 (5′-catatggctgctgaagat-3′) and oligonucleotide 4 (5′-aagcttaaccctcatcatcact-3′) as the 5′ and 3′ primers, respectively. The PCR fragment was digested with NdeI–HindIII and subcloned into NdeI–HindIII-cut pET28a. This construct allows the expression in E. coli of RanGAP1 as a fusion protein with an N-terminal 20-aa, including a 6-aa His tag. The resultant plasmid was transformed into E. coli [strain BL21 (DE3) pLysS], and RanGAP1 expression was induced under standard conditions (16). The His-tagged RanGAP1 was purified on a Ni+2-agarose column (Novagen). The protein was further purified by excising the RanGAP1 band after SDS/PAGE. The gel slice containing RanGAP1 was used to elicit antibody production in two rabbits (Research Genetics). Purified preparations of His-tagged RanGAP1 were coupled to CNBr-activated Sepharose (Pharmacia), and used for the production of affinity-purified antibodies following standard methods (23).
Guinea pig anti-p340RanBP2 antibodies were as described in ref. 16. Horseradish-peroxidase-conjugated anti-rabbit or anti-guinea pig IgG antibodies were purchased from Amersham or Jackson ImmunoResearch.
Immunoprecipitation.
Egg extract (100 μl) was incubated with 1 μl of antiserum for 30 min at room temperature. Buffer A (400 μl) and 20 μl of protein A-Sepharose beads (Pharmacia) were added. After incubation for 1 hr at 4°C, the beads were collected by centrifugation and washed three times in buffer A. The proteins coprecipitated with the beads were eluted by boiling in SDS-sample buffer. After separation by SDS/PAGE, proteins were visualized by silver staining or subjected to Western blot analysis (23).
Gel Filtration Column Chromatography.
Egg extract (50 μl) was diluted with 50 μl of column buffer, and insoluble material was removed by centrifugation for 5 min in an Eppendorf centrifuge. The mixture was loaded onto a Sephacryl S300 gel filtration column (1.0 × 30 cm; Pharmacia). Column chromatography was carried out at 4°C at a flow rate of 200 μl/min in column buffer. Aliquots (10 μl) of each fraction were analyzed by SDS/PAGE followed by Western blot analysis. Protein size markers were purchased from Bio-Rad.
RanGAP Assay Using Recombinant Ran.
Purified recombinant human Ran (2 μM) and 20 nM [γ-32P]GTP (6,000 Ci/mmol; 1 Ci = 37 GBq; Dupont/NEN) or [α-32P]GTP (3,000 Ci/mmol; Dupont/NEN) were incubated for 10 min at 25°C in 50 μl of buffer containing 20 mM Tris·HCl, pH 8.0/50 mM NaCl/10 mM EDTA. The buffer was changed to GAP buffer (20 mM Tris·HCl, pH 8.0/50 mM NaCl/5 mM MgCl2/0.05% BSA) on a NAP-5 column (Pharmacia). To measure RanGAP activity, immunoprecipitated beads were incubated in 100 μl of GAP buffer containing 100 nM radiolabeled Ran at 25°C for the indicated times. Each reaction was filtered through nitrocellulose BA85, 0.45 μm (Schleicher & Schuell), followed by three washes with 1 ml of GAP buffer. The radioactivity on the filters was counted using LS-5000-TD scintillation counter (Beckman).
RESULTS
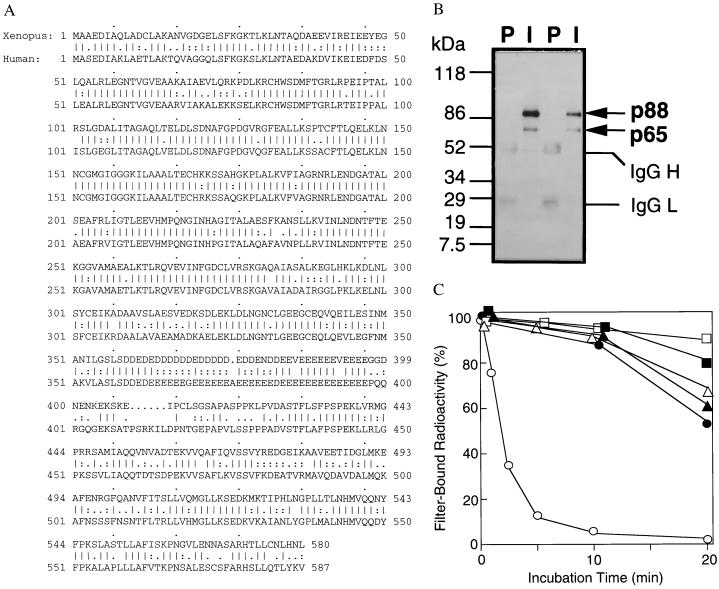
We previously reported that Xenopus RanBP2/Nup358 associates tightly with at least two other peptides, p88 and p18 (ref. 16; Fig. 1). Analysis of peptides derived from p88 indicated that it is either a modified form of the Ran GAP, RanGAP1/Fug1, or a very closely related protein (16). To determine the identity of p88, we cloned the Xenopus homologue of RanGAP1 (see Materials and Methods) and examined its behavior in egg extracts. The clone encoded a peptide with an ORF of 580 aa (Fig. 2A). Alignment of the encoded peptide with human and murine RanGAP1 (20, 24) reveals a high level of protein sequence homology (69% identity, 84% similarity). We expressed the N-terminal 360 residues of RanGAP1 in E. coli, purified this fragment, and used it for generating polyclonal antibodies in rabbits. These anti-RanGAP1 antibodies recognized two bands on Western blots of Xenopus extracts. One band migrated with an apparent molecular mass of 65 kDa, similar to the migration of mammalian RanGAP1 proteins (20, 24) and close to the predicted molecular mass of Xenopus RanGAP1 (63.9 kDa). The other band comigrated with p88, the RanGAP1-like protein we had observed in association with p340RanBP2. To determine whether the upper form of RanGAP1 might be p88, we performed Western blot analysis of anti-p340RanBP2 immunoprecipitates with the anti-RanGAP1 antibodies and found that p88 was indeed the upper band recognized by the anti-RanGAP1 antibody (Fig. 2B; see Fig. 4).
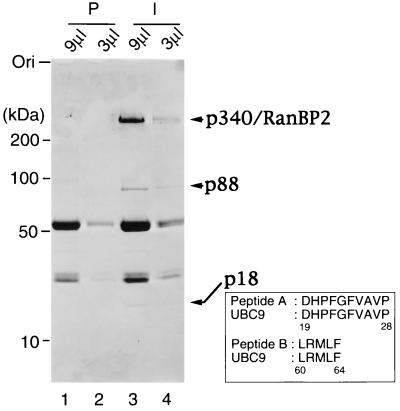
Figure 1.
p18UBC9 and p88RanGAP1 are precipitated by anti-p340RanBP2 antibodies. Egg extract (100 μl) was incubated with preimmune serum (lanes 1 and 2) or with anti-p340RanBP2 serum, followed by precipitation with protein A-Sepharose beads. The beads were washed, and the bound proteins were eluted with 20 μl of SDS-sample buffer. Either 9 μl (lanes 1 and 3) or 3 μl (lanes 2 and 4) of eluted proteins was subjected to SDS/PAGE and Coomassie blue staining. The migration of molecular mass standards (in kDa) is indicated on the left. Arrows on the right indicate proteins that were specifically immunoprecipitated by the anti-p340RanBP2 antibodies. Two peptide sequences derived from p18 are shown as Peptides A and B, with the corresponding sequences from mammalian Ubc9p indicated below each.
Figure 2.
p88 is a modified form of RanGAP1. (A) Protein sequence of Xenopus RanGAP1. The protein sequence of human RanGAP1 is shown below for comparison. Straight vertical lines indicate identical amino acids. Double dots indicate conserved residues, and single dots indicate charged residues. Gaps inserted for sequence alignment are indicated by periods. (B) p88 is a modified form of RanGAP1. Immunoprecipitations were performed from egg extracts using affinity-purified anti-RanGAP1 antibodies (I) or preimmune sera (P) from two rabbits. The precipitated proteins were analyzed by Western blotting with affinity-purified anti-RanGAP1 antibodies from one rabbit. The migration of molecular mass standards (in kDa) is indicated on the left. The two RanGAP1 bands (p88 and p65) and the IgG H and L chains are indicated on the right. (C) p340RanBP2–p88–p18 complexes contain RanGAP activity. Beads (1 μl) from an anti-p340RanBP2 immunoprecipitation (○ and •), a preimmune control incubation (▵ and ▴), or an incubation with protein A-Sepharose lacking IgG (□ and ▪) were added to GAP buffer containing 100 nM Ran–[γ-32P]GTP (○, ▵, and □) or 100 nM Ran–[α-32P]GTP (•, ▴, and ▪). At the indicated times, protein-associated 32P was measured in a filter binding assay.
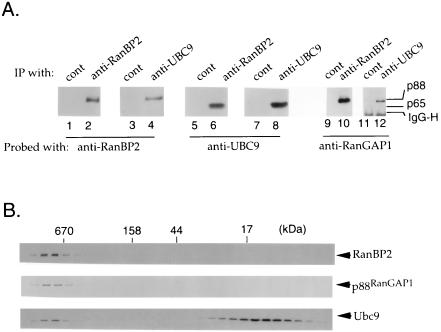
Figure 4.
The distribution of p340RanBP2, p88, and p18 in Xenopus egg extracts. (A) Immunoprecipitation experiments were carried out using anti-p340RanBP2 serum (lanes 2, 6, and 10) or anti-Ubc9p serum (lanes 4, 8, and 12). Control experiments were performed with corresponding guinea pig (lanes 1, 5, and 9) and rabbit (lanes 3, 7, and 11) preimmune sera. Immunoprecipitated proteins were subjected to Western blot analysis with anti-p340RanBP2 (lanes 1–4), anti-Ubc9p (lanes 5–8), or anti-RanGAP1 (lanes 9–12) serum. The positions of the 88- and 65-kDa forms of RanGAP1 are indicated to the right of lane 12. The Ig H chain detected in the Western blotting lanes 11 and 12 is indicated as IgG-H. (B) Egg extract (50 μl) was gel-filtered by Sephacryl S300 column chromatography. Fractions were analyzed by Western blotting using anti-p340RanBP2 (Top), anti-RanGAP1 (Middle), or anti-Ubc9p (Bottom) serum.
When we expressed the full-length RanGAP1 clone in bacteria, a single protein was produced, and it migrated with an apparent molecular mass of 65 kDa on SDS/PAGE gels. This band precisely comigrated with the smaller form of RanGAP1 found in Xenopus extracts. However, when we expressed this RanGAP1 clone in a reticulocyte lysate-coupled transcription–translation system, we observed two labeled bands that migrated with apparent molecular masses of 65 and 88 kDa, matching those of the two bands detected by Western blotting of Xenopus extracts (data not shown). Further, bacterially expressed, tagged RanGAP1 became modified when added to Xenopus egg extracts and showed a shift in its electrophoretic mobility corresponding to an increase in apparent molecular mass of approximately 23 kDa (data not shown). Taken together, these results argue that p88 is a modified form of RanGAP1, and that the factors required for performing this modification are present in both Xenopus egg extracts and reticulocyte lysates. Western blot analysis confirms that both forms of RanGAP1 are also present in Xenopus tissue culture cells and in intact oocytes and eggs (data not shown). We have never observed the 65-kDa form of RanGAP1 associated with p340RanBP2 despite repeated attempts (see Fig. 4), suggesting either that modification may be required before association with p340RanBP2 or that association with p340RanBP2 may facilitate modification.
The results presented above are consistent with the idea that GTP hydrolysis by Ran facilitates protein import in association with the p340RanBP2–p88–p18 complex. To test this idea further, we wished to determine whether p88 is an active RanGAP. To do this, we examined whether the proteins associated with anti-p340RanBP2 immunoprecipitates have significant GAP activity by measuring the release of 32P from [γ-32P]GTP-bound Ran protein in a filter binding assay (Fig. 2C). We observed that the anti-p340RanBP2 immunoprecipitates activated GTP hydrolysis by Ran. Control beads that were similarly prepared using preimmune serum rather than immune serum did not significantly promote GTP–Ran hydrolysis in this assay. We quantitated the amount of p88 in the reactions by comparison of Coomassie blue-stained bands of p88 on SDS/PAGE gels to concentration standards. On the basis of this quantitation, we estimate that p88 has a specific activity as a Ran GAP in the same order of magnitude as that estimated for unmodified form of mammalian RanGAP1 (data not shown; ref. 20). These data argue that the modification of RanGAP1 does not result in its inactivation as a GAP. Rather, modification may alter its localization and/or its association with p340RanBP2 or other proteins. It is also possible that modification might alter the stability of RanGAP1, although neither form of RanGAP1 appeared to be particularly unstable in our hands.
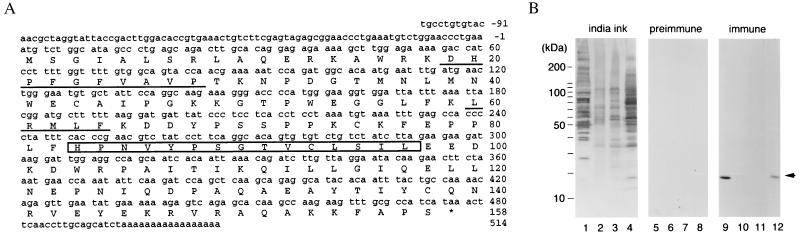
We wished to identify p18, and to determine whether it might have a role in regulating the activity of p340RanBP2 or p88. To identify p18, we purified several micrograms through immunoprecipitation using anti-p340RanBP2 antibodies. This protein was subjected to peptide sequence analysis, from which we obtained two peptide sequences (Fig. 1). Both sequences were identical to sequences in mammalian Ubc9p (12–15). We cloned a cDNA encoding p18 from Xenopus oocyte cDNA by PCR (Fig. 3A) and found that it showed 100% sequence identity with human UBC9 at the protein level. A His-tagged version of Ubc9p was expressed in E. coli, purified by affinity chromatography on a Ni+2-agarose column, and used to generate antibodies in rabbits (Fig. 3B). These sera from rabbits recognized a single band of 18 kDa in Western blot analysis of Xenopus egg extracts. In fractionated extracts (22), this protein was found almost exclusively in the soluble fraction and not found in the membrane fraction. This distribution is similar to that observed for Ran and p340RanBP2 (16).
Figure 3.
Cloning of Xenopus UBC9 and preparation of anti-Ubc9p antibodies. (A) Nucleotide and predicted amino acid sequence of Xenopus laevis UBC9 cDNA. The amino acid sequences obtained by peptide sequencing of p18 are underlined. The E2 active site motif, HPN(I/V)X3–4GX(I/V/L)C(I/L)X(I/V)(I/L) is boxed. (B) Xenopus egg cytosolic fraction (1 μl; lanes 1, 5, and 9), 0.1 μl of membrane faction (lanes 2, 6, and 10), or 0.1 μl of glycogen pellet fraction (lanes 3, 7, and 11) were blotted to poly(vinylidene difluoride) membranes for analysis. On the same membranes, 30 μg of total protein from Xenopus A6 tissue culture cells was also blotted (lanes 4, 8, and 12). One membrane was stained with India ink to show the transfer of proteins to the membrane (lanes 1–4). The other filters were subjected to Western blot analysis using preimmune sera (lanes 5–8) or rabbit polyclonal antiserum prepared against recombinant Xenopus Ubc9p (lanes 9–12) at a dilution of 1:5,000. The migration of molecular mass standards (in kDa) are indicated to the left in A and B. Egg interphase cytosol, membrane, and glycogen-rich fractions were prepared as described in Smythe and Newport (22). The arrow at the right indicates the position of p18.
To confirm the association of p340RanBP2, p88, and p18, we performed immunoprecipitation experiments using the antisera against p340RanBP2 and Ubc9p. The proteins associated with both immunoprecipitations were analyzed by Western blotting with anti-p340RanBP2, anti-RanGAP1, and anti-Ubc9p antisera (Fig. 4A). As predicted from our earlier experiments, we found that antibodies directed against either protein precipitated the other two. We did not observe any of the 65-kDa form of RanGAP1 associated with anti-p340RanBP2 or anti-Ubc9p immunoprecipitates (Fig. 4A), nor do we find this form of RanGAP1 as a major protein removed from extracts in association with a glutathione S-transferase (GST)–Ran fusion protein (16). The results of these experiments support the existence of complexes containing all three proteins and confirm the identity of Ubc9p as the protein we had previously observed in association with p340RanBP2 and GST–Ran. Despite the association of p18 with the anti-p340RanBP2 immunoprecipitates, we did not find that any of the proteins associated with p340RanBP2 were ubiquitinated, as judged by Western blotting with anti-ubiquitin antibodies (data not shown).
We wished to determine the distribution of p88 and p18 between their p340RanBP2-associated and free pools. The experiments presented in Fig. 4A are consistent with the existence of a complex containing all three proteins, but they were not informative regarding what percentage of the total pools of each protein was in such complexes. To address this question, we determined whether the three proteins cofractionated in gel filtration analysis of Xenopus egg extracts using Sepharose S300 (Fig. 4B). In this analysis, all the p340RanBP2 migrated at a very high molecular mass, as might be anticipated from its size. The majority of p88 cofractionated with p340RanBP2 (Fig. 4B), but the unmodified RanGAP1 eluted in a broad peak centered at an apparent molecular mass of approximately 150 kDa (data not shown). The elution of unmodified RanGAP1 in these column fractions was consistent with previous observations that unmodified mammalian RanGAP1 forms a homodimer (25). The majority of p18 behaved as if it were an unbound protein or associated only with other proteins of a low molecular mass. However, roughly 10% of the p18 protein migrated in a distinct peak that cofractionated with p340RanBP2. A small portion of the total Ran pools also cofractionated with p340RanBP2 (16), so it is possible that it was associated with the p340–p88–p18 complex. However, we do not believe that Ran is required for the association of these proteins because our previous experiments have indicated that complex formation is independent of the nucleotide binding state of Ran (16). This analysis further supports the existence of a p340–p88–p18 complex and demonstrates that a significant fraction of each of the three components cofractionates from egg extract, substantiating the idea that the observed complex represents specific association of the three proteins.
DISCUSSION
In this report, we have presented evidence that the Xenopus homologues of RanBP2, RanGAP1, and Ubc9p form a tight complex in egg extracts. We have also documented that RanGAP1 is posttranslationally modified in this complex. This modification was linked with the assembly of the complex because we did not find any unmodified RanGAP1 in association with p340RanBP2 or p18UBC9. The modification of RanGAP1 did not abolish its activity as a GAP for Ran. It has previously been suggested that RanBP2 may coordinate the assembly of protein machines required for translocation of docked import substrates into the nucleus (19), and the association of p88RanGAP1 with p340RanBP2 supports this notion. The association of p18UBC9 with p340RanBP2 suggests that that Ubc9p may either regulate transport by modification of pore proteins or modify nuclear proteins in a transport-coupled manner.
Experiments designed to test whether p88RanGAP1 is a ubiquitinated or phosphorylated form of p65RanGAP1 yielded negative results. Anti-ubiquitin antibodies did not recognize p88 in Western blot analysis, and p88 was not converted to p65 by phosphatase treatment (data not shown). Although such negative results are not conclusive, we believe that p65 is converted to p88 by a different modification. After this manuscript was submitted, two reports indicating that mammalian RanGAP1 is converted from a 70-kDa form to a 90-kDa form by conjugation with a small ubiquitin-like protein were published (26, 27). This ubiquitin-like protein has been named UBL1 (28), PIC1 (29), SUMO-1 (26), or GMP1 (27). (Here we will call it UBL1 as no consensus has been established regarding the nomenclature for it.) Our preliminary data strongly suggest that the modification of Xenopus RanGAP1 that converts it from the 65-kDa form to the 88-kDa form is also conjugation with UBL1 (data not shown). In this report, we have presented evidence that modification of RanGAP1 regulates its association with p340RanBP2 (Fig. 4) rather than drastically altering its activity or stability. Consistent with this conclusion, Mahajan et al. (26) recently demonstrated that conjugation of mammalian RanGAP1 with UBL1 promotes its interaction with RanBP2/Nup358. The association of modified RanGAP1 with p340RanBP2 could result from direct recognition of the modified domain of RanGAP1, or from an alteration in RanGAP1’s own conformation or dimerization state.
In S. cerevisiae, repression of Ubc9p synthesis blocks cell cycle progression in late G2 or early M phase, and results in the stabilization of B-type cyclins (9). This phenotype is strikingly similar to the phenotype of srp1 mutants, which also arrest in G2/M phase and which cannot degrade B-type cyclins during G1 phase when they are normally unstable (30). The srp1 gene encodes the yeast homologue of the nuclear localization signal sequence receptor/importin α protein, which targets karyophilic proteins to docking sites on the nuclear pore before their translocation into the nucleus (1). It is tempting to speculate that the transport of a mitotic regulatory factor(s) into the nucleus may be critical both for the onset of mitosis and for the destruction of mitotic cyclins, and that Ubc9p might promote mitosis by modifying RanGAP1 or other components of transport machinery to augment the import of this factor. Our preliminary results are consistent with the idea that Ubc9p acts as an E2 enzyme for the conjugation of UBL1 with other proteins, but we have not yet obtained definitive results proving that this is the case. This idea is also circumstantially supported by reports that Ubc9p associates with UBL1 in other systems (28). Alternatively, Ubc9p might regulate mitosis by modifying mitotic factors themselves in a transport-coupled manner. Ubc9p associates with a number of nuclear proteins (12–15), and it will be of interest to determine whether these proteins might be targets for transport-coupled modification by Ubc9p.
We originally observed the p340–p88–p18 complex in association with GST–Ran (16). There is a considerable body of evidence indicating that the Ran GTPase pathway is required for the proper regulation of mitosis with respect to the completion of DNA replication (6). We would finally note the possibility that UBC9 mutants and mutants in the Ran GTPase pathway may both be unable to properly control the G2- to M-phase transition because they may both result in misregulated subcellular localization of mitotic factors. In the future, it will be important to determine whether this is true, and if it is, to understand the point at which these proteins are required for correct regulation of the cell cycle.
Acknowledgments
We would like to thank Michael Herrler for his help in cloning the cDNA for Xenopus UBC9. We would like to thank Heather Shelsta and Marvin Carr for their help in sequencing the Xenopus RanGAP1 clone. We also thank Alan Weissman and Jane Jensen for assistance with Western blot analysis using anti-ubiquitin antibodies.
ABBREVIATION
- GAP
GTPase-activating protein
Footnotes
References
- 1.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 2.Rush M G, Drivas G, D’Eustachio P. BioEssays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- 3.Sazer S. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- 4.Nishimoto T, Eilen E, Basilico C. Cell. 1978;15:475–483. doi: 10.1016/0092-8674(78)90017-x. [DOI] [PubMed] [Google Scholar]
- 5.Nishitani H, Ohtsubo M, Yamashita K, Iida H, Pines J, Yasuda H, Shibata Y, Hunter T, Nishimoto T. EMBO J. 1991;10:1555–1564. doi: 10.1002/j.1460-2075.1991.tb07675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasso M. Prog Cell Cycle Res. 1995;1:163–172. doi: 10.1007/978-1-4615-1809-9_13. [DOI] [PubMed] [Google Scholar]
- 7.Kornbluth S, Dasso M, Newport J W. J Cell Biol. 1994;125:705–720. doi: 10.1083/jcb.125.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke P R, Klebe C, Wittinghofer A, Karsenti E. J Cell Sci. 1995;108:1217–1225. doi: 10.1242/jcs.108.3.1217. [DOI] [PubMed] [Google Scholar]
- 9.Seufert W, Futcher B, Jentsch S. Nature (London) 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 11.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 12.Kovalenko O V, Plug A W, Haaf T, Gonda D K, Ashley T, Ward D C, Radding C M, Golub E I. Proc Natl Acad Sci USA. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Koltin Y. Mol Gen Genet. 1996;251:153–160. doi: 10.1007/BF02172913. [DOI] [PubMed] [Google Scholar]
- 14.Yasugi T, Howley P M. Nucleic Acids Res. 1996;24:2005–2010. doi: 10.1093/nar/24.11.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hateboer G, Hijmans E M, Nooij J B D, Schlenker S, Jentsch S, Bernards R. J Biol Chem. 1996;271:25906–25911. doi: 10.1074/jbc.271.42.25906. [DOI] [PubMed] [Google Scholar]
- 16.Saitoh H, Cooke C A, Burgess W H, Earnshaw W C, Dasso M. Mol Biol Cell. 1996;7:1319–1334. doi: 10.1091/mbc.7.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. Nature (London) 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 19.Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester W, Stutz F, Rosbash M, Wickens M. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 22.Smythe C, Newport J W. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 24.DeGregori J, Russ A, von Melchner H, Rayburn H, Priyaranjan P, Jenkins N A, Copeland N G, Ruley H E. Genes Dev. 1994;8:265–276. doi: 10.1101/gad.8.3.265. [DOI] [PubMed] [Google Scholar]
- 25.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 27.Matinus M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 29.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 30.Loeb J D, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G R. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]