Abstract
Mitogen-activated protein kinase kinase (MEK) is a dual-specificity protein kinase that is located primarily in the cellular cytosol, both prior to and upon mitogenic stimulation. The existence of a nuclear export signal in the N-terminal domain of MEK [Fukuda, M., Gotoh, I., Gotoh, Y. & Nishida, E. (1996) J. Biol. Chem. 271, 20024–20028] suggests that there are circumstances under which MEK enters the nucleus and must be exported. Using mutants of MEK, we show that the deletion of the nuclear export signal sequence from constitutively active MEK caused constitutive localization of MEK in the nucleus of COS7 and HEK-293T cells. However, when the same region was deleted from a catalytically inactive MEK, cytoplasmic localization was observed in resting cells, which turned nuclear upon stimulation. Confocal microscopy of COS7 cells expressing the above mutants showed localization of the active MEK in the nuclear envelope and also in the cell periphery. The differences in cellular localization between the wild-type and mutant MEKs are not due to severe changes in specificity because the recombinant, constitutively active MEK that lacked its N-terminal region exhibited the same substrate specificity as the wild-type MEK, both in vitro and in intact cells. Taken together, our results indicate that upon mitogenic stimulation, MEK, like extracellular signal responsive kinase and p90RSK, is massively translocated to the nucleus. Rapid export from the nucleus, which is mediated by the nuclear export signal, is probably the cause for the cytoplasmic distribution observed with wild-type MEK.
Keywords: signal transduction, substrate specificity
The transmission of extracellular signals from the cell surface into the nucleus involves several groups of protein kinases, which are collectively known as the mitogen-activated protein kinase (MAPK) signaling cascades. One of these cascades, the extracellular signal responsive kinase (ERK) signaling cascade, involves a sequential phosphorylation and activation of Raf1, MAPK kinase (MAPKK, also known as MEK), ERK, p90RSK, and under some conditions also GSK3 (reviewed in ref. 1). Other MAPK signaling cascades are the JNK (stress-activated protein kinases) cascade, the p38RK (HOG, reviewed in ref. 2), and other, less-characterized cascades. A key step in the signaling mechanism of the ERK cascade is the translocation of both ERK1 and 2 and p90RSK into the nucleus (3, 4). This translocation, which occurs in response to mitogenic stimulation, is rapid (occurs within 5–30 min), and might be a prerequisite for activation of nuclear processes such as transcription (5, 6). The mechanism by which protein kinases are translocated to the nucleus upon stimulation is not yet known. The sequences of the ERKs and p90RSK do not contain a nuclear localization signal (NLS), and although the size of ERK may permit a simple diffusion via nuclear pores, such diffusion would probably be much slower than the rapid movement observed upon activation. However, because kinase-deficient mutants of ERK can translocate to the nucleus (4), clearly the mechanism of its nuclear translocation is distinct from the mechanism of activation. In contrast to ERK and p90RSK, their upstream regulator, MEK is absent from the nucleus both prior to and upon extracellular stimulation (4, 7). The cytoplasmatic localization of MEK is homogeneous and, unlike ERKs, it does not seem to associate with cytoskeletal elements (4, 8). This subcellular distribution might be important for the activation of MEK by its membrane-associated, upstream activator, Raf1, and recently this localization was attributed to a segment in the MEK’s N-terminal region (9) that acts as a nuclear export signal (NES; ref. 10 and 11).
The existence of an NES sequence in the cytoplasmic MEK raises the question: What are the circumstances under which MEK enters the nucleus and has to be exported? In this manuscript, we present evidence that, upon mitogenic stimulation, MEK, like ERK, is massively translocated to the nucleus. Rapid export from the nucleus, which is mediated by the NES, probably accounts for the apparent cytoplasmic distribution observed in the previous reports. We also report accumulation of active MEK in the cell periphery and in the nuclear envelope, which indicates that MEK might also regulate processes in these locations.
MATERIALS AND METHODS
MEK Constructs.
The following MEK mutants were used: WT-MEK and KA-MEK, wild-type and catalytically inactive MEK1, were prepared as previously described (12, 13); EE-MEK was prepared by substituting serines 218 and 222 in MEK1 with glutamic acids (14, 15); for ΔN-EE-MEK and ΔN-MEK, amino acids 32–51 were deleted from EE- or WT-MEKs by removing the StuI 95–154 fragment (16); and ΔN-KA-EE-MEK was obtained by replacing the StuI 155–527 fragment from ΔN-EE-MEK with the same fragment from KA-MEK. These constructs were introduced either into the expression vector pCDNA1 for transient expression (12) or into pRSETa (both from Invitrogen) to express N-terminal 6 histidine (6His)-linked MEK constructs in bacteria.
Expression of MEK. Transient expression.
COS7 monkey kidney cells and 293T modified human embryonic kidney fibroblasts (HEK-293T) were grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (FCS; GIBCO/BRL) as previously described (12). COS7 and HEK-293T (50–70% confluent) were transfected by the DEAE-dextran (17) or calcium phosphate (18) methods, respectively.
Expression in bacteria.
6His-ΔN-EE- and 6His-WT-MEKs in pRSETa were introduced into BL21 bacteria. These bacteria were grown until OD600 = 0.7–1.0 and isopropyl-β-d-thio-galactopyranoside (ChemBridge, Northbrook, IL, 0.4 mM) was added for 4 hr. The 6His-MEKs were purified on a nickel column (Probond, Invitrogen) according to the manufacturer’s instructions. The proteins, which were eluted with 0.1 M imidazole, were 75% pure and kept at 4°C for up to 6 weeks.
Cell Staining.
One day after transfection, the cells were placed on glass coverslips. After growing for an additional day, the cells were serum-starved in 0.1% FCS (8). Stimulation, fixation (3% paraformaldehyde), permeabilization (0.2% Triton X-100, 5 min), and staining were as described (8). The cells were sequentially incubated (45 min each) with monoclonal anti-MEK1 antibodies (1:30, Transduction Laboratories, Lexington, KY) and rhodamine-conjugated goat–anti-mouse antibodies (1:200, Jackson ImmunoResearch).
Coexpression and Activity Assays of MAPKs.
COS7 cells in 10-cm plates were cotransfected with 10 μg of ΔN-EE-MEK in pcDNA1, and 2 μg of hemagglutinin (HA) fragment-tagged ERK2, p38RK, or JNK2 in the plasmid SRα. One day later, each plate was divided into two, kept growing for an additional 24 hr, and serum-starved as above. One plate of each pair was stimulated and the other served as a basal control. After stimulation, the cells were washed, lysed, and immunoprecipitated with monoclonal anti-HA (Antibodies Unit, The Weizmann Institute of Science) using protein A-agarose (20 μl, Sigma), as previously described (19). During the final step of immunoprecipitation, ERK and p38RK were washed with buffer A (50 mM β-glycerophosphate, pH 7.3/1.5 mM EGTA/1 mM EDTA/1 mM DTT/0.1 mM Na3VO4), resuspended in 15 μl of buffer A, and incubated (20 min, 30°C) with 5 μl of 2 mg/ml myelin basic protein (MBP) and 10 μl of 3× reaction mix [30 mM MgCl2/4.5 mM DTT/75 mM β-glycerophosphate, pH 7.3/0.15 mM Na3VO4/3.75 mM EGTA/30 μM calmidazolium/2.5 mg/ml BSA/100 μM [γ32P]-ATP (2 cpm/fmol; ref. 13)]. JNK precipitates were washed with kinase buffer (20 mM Hepes, pH 7.7/2 mM DTT/20 mM MgCl2/40 mM β-glycerophosphate/0.1 mM Na3VO4) and resuspended in 40 μl of the same buffer containing GST-Jun 1–97 (10 μl, 3 μg) and [γ32P]-ATP (20). All reactions were terminated by sample buffer and analyzed by 10% SDS/PAGE.
Cellular Fractionation.
Nuclear and cytosolic fractions were separated as previously described (21). Briefly, after stimulation, the cells (3 × 106) were washed, scraped into 0.5 ml of buffer A supplemented with protease inhibitors (13), centrifuged (12,000 × g, 5 min, 4°C), and resuspended in 200 μl of the lysis buffer containing 0.1% Nonidet P-40. The lysate was mixed vigorously and centrifuged as above to yield a supernatant that contained the cytosol fraction. Nuclear proteins were extracted by resuspending the nuclear pellet in 100 μl of extraction buffer (420 mM NaCl/50 mM β-glycerophosphate/0.5 mM Na3VO4/1.5 mM MgCl2/0.2 mM EDTA/1 mM DTT/25% glycerol), brief sonication (2 × 5 sec, 40 W, 4°C), vigorous mixing, and centrifugation. The purity of this fraction was >85%, as judged by anti-tubulin antibodies.
MAPKK Activity.
MAPKK (MEK1 and MEK2) activity was measured, after partial purification on DE52 minicolumns, by the net activation of recombinant ERK2 toward MBP as previously described (13).
Phosphorylation in Vitro.
Phosphorylation of proteins and peptides by MEK was done as previously described (22) in the reaction mixture used for MBP phosphorylation as above.
RESULTS
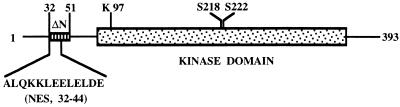
The existence of a sequence similar to the nuclear export signal in the N-terminal region of MEK (amino acids 32–44) raised the possibility that MEK is translocated to the nucleus upon stimulation. To study this hypothesis, we first deleted the N-terminal stretch (amino acids 32–51) that contains the NES region from both the constitutively active MEK (EE-MEK; refs. 14 and 15) and MEK1, thus generating ΔN-EE- and ΔN-MEKs, respectively. Similar constructs were previously reported to express high (ΔN-EE-MEK) or moderate (ΔN-MEK) constitutive MAPKK activity (16). The two properties of MEK (activity and localization) that might be determined by this N-terminal region were distinguished by the substitution of lysine 97 to alanine in ΔN-EE-MEK (ΔN-KA-EE-MEK), resulting in a catalytically inactive enzyme lacking its NES (Fig. 1).
Figure 1.
Schematic representation of MEK1. The sites of mutations are indicated.
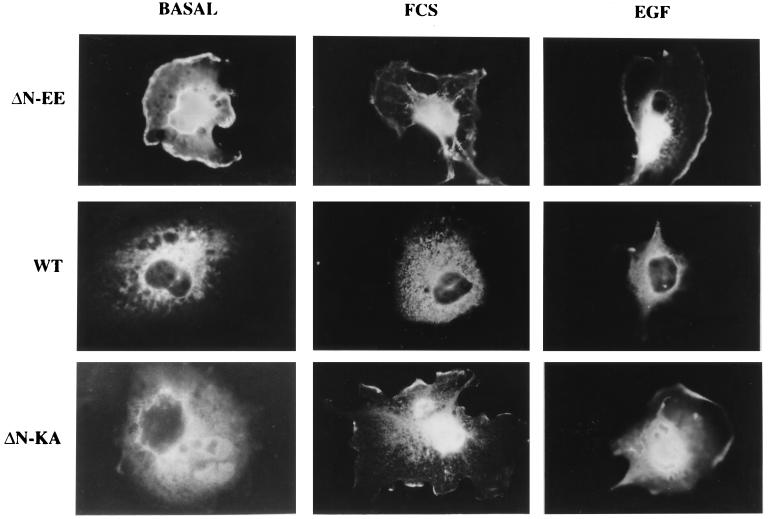
COS7 monkey kidney cells were transfected with the above constructs, and then fixed and stained with monoclonal anti-MEK antibodies (Fig. 2). In serum-starved COS7 cells transfected with ΔN-EE-MEK, staining was observed primarily in the nucleus (>50%). Significant staining was also detected in the cytoplasm and in the cell periphery (possibly lamelopodia) and was probably specific (not edge effect), because it was not detected in other overexpressing cells. As expected under these conditions, WT-MEK was distributed homogeneously in the cytosol and completely absent from the nucleus. Serum stimulation of the cells overexpressing both constructs, which caused a substantial shift of ERK to the nucleus (≈80%, data not shown), resulted in only a small increase (5–20%) in the nuclear staining of MEK. Stimulation of the same cells with epidermal growth factor (Fig. 2), or with phorbol 12-myristate 13-acetate (PMA; not shown), which caused 40% translocation of ERK, induced small nuclear shift similar to the serum treatment. ΔN-MEK gave a similar trend of results as ΔN-EE-MEK (not shown). A different picture, however, was obtained in cells overexpressing the inactive construct, ΔN-KA-EE-MEK. In resting cells, the mutant protein was detected only in the cytoplasm, whereas upon FCS, epidermal growth factor, or PMA stimulation, the majority (>75%) of the ΔN-KA-EE-MEK protein was detected in the nucleus, with slight staining in the cell periphery (as for ΔN-EE-MEK). Staining of endogenous MEK could not account for the observed results, since positive staining was not observed in vector control cells subjected to the same conditions.
Figure 2.
Localization of the overexpressed ΔN-EE-, WT-, and ΔN-KA-EE (ΔN-KA)- MEKs in COS7 cells. COS7 cells transfected with each one of these constructs were subjected to serum starvation for 16 hr and then either stimulated with 10% FCS (10 min), or epidermal growth factor (EGF; 50 μg/ml, 5 min), or left untreated (basal). The cells were stained with anti-MEK1 antibodies and visualized using conventional fluorescent microscopy (Zeiss microscope magnification of ×800).
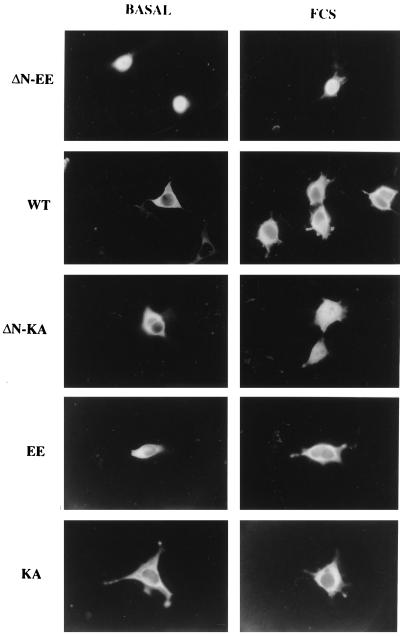
To verify that the observed staining was indeed a general phenomenon and not due to high overexpression of the MEK mutants in COS7 cells, the above transfections were repeated with the human embryonic kidney cell line (HEK-293T). The localization of staining of ΔN-EE-, WT-, or ΔN-KA-EE-MEKs in these cells was similar to that observed in COS7 cells, except for the absence of clear staining in the cell periphery of the HEK-293T cells (Fig. 3). In resting cells, ΔN-EE-MEK was localized primarily in the nucleus with some slight staining in the cytoplasm, and this staining was not changed upon stimulation. WT-MEK was localized in the cytosol of the HEK-293T cells before stimulation, and a small, but detectable, shift of the kinase to the nucleus occurred after stimulation. With ΔN-KA-EE-MEK, staining was localized in the cytoplasm before stimulation and most of it shifted to the nucleus after addition of serum. For controls, EE-MEK and KA-MEK were overexpressed in the HEK-293T cells. The constitutively active EE-MEK, which still contains its NES, was located chiefly in the cytoplasm, although some staining (<25%) could be detected in the nucleus and was not affected by serum stimulation; KA-MEK was observed mainly in the cytosol under all the conditions tested (Fig. 3). A similar staining trend with the EE- and KA-MEKs was also observed in overexpressing COS7 cells (not shown). It should be noted that some COS7 cells that overexpressed ΔN-KA-EE-MEK displayed nuclear localization even after serum starvation, which increased after stimulation. This uncharacteristic staining could result from incomplete starvation of the cells, a suggestion supported by the finding of ERK in the nuclei of some vector transfected control cells (not shown).
Figure 3.
Localization of overexpressed ΔN-EE-, WT-, ΔN-KA-EE-, EE-, and KA-MEKs in HEK-293T cells. Staining was assessed by conventional fluorescent microscopy as in Fig. 2.
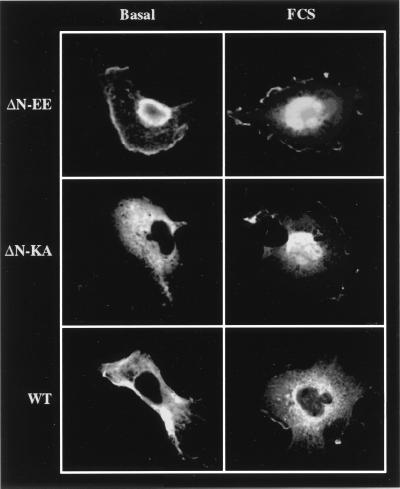
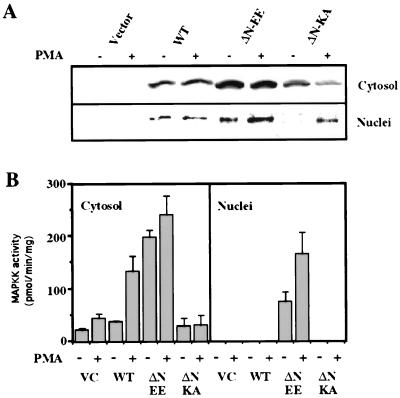
Because some MEK mutants showed an unequal distribution of staining, confocal microscopy was used to identify the exact sublocalization of the various mutants. In resting cells, the nuclear staining of ΔN-EE-MEK seemed to be localized mainly in the periphery, with little staining detected in the center of this organelle (Fig. 4). In these cells, the rest of the staining was dispersed in the cytoplasm or attached to the periphery of the cells. Upon stimulation, the staining of the nucleus became more homogenous, although the staining in the center was still low. The staining observed with ΔN-KA-EE- and WT-MEKs was similar to that described in Fig. 2, except that upon the stimulation, WT-MEK showed low nuclear staining localized in defined structures in the center. The nuclear distribution of the various MEK mutants was further verified by fractionation of subcellular compartments. Western blotting (Fig. 5A) of purified nuclei from COS7 cells revealed a distribution of the expressed mutants similar to that of the staining experiments. Assays of enzymatic activity (Fig. 5B) showed that WT-MEK had a low basal activity in the cytosol that was elevated upon PMA stimulation, but no activity could be detected in the nucleus. MAPKK activity in cells expressing ΔN-EE-MEK was high both in the cytoplasm and in the nucleus, whereas the activity in the ΔN-KA-EE-MEK cells was low in the cytosol and absent from the nucleus. Taken together, these results corroborate those of the staining experiments.
Figure 4.
Subcellular localization of ΔN-EE-, WT-, and ΔN-KA-EE-MEKs in COS7 cells by confocal microscopy. The slides used in Fig. 2 were viewed by confocal microscope. (Bio-Rad MRC-1024, objective ×20, zoom 3.6.)
Figure 5.
Determination of the cellular localization of ΔN-EE-, WT-, and ΔN-KA-EE-MEKs in COS7 cells using fractionation. COS7 cells overexpressing the constructs were either left untreated (basal control), or stimulated with PMA (250 nM, 10 min). The cells were fractionated and subjected to (A) Western blotting with monoclonal anti-MEK1 antibodies (due to differences in amounts of overexpression, the amount of MEK in this figure can only be compared in the basal and stimulated pairs that were derived from the same transfection), or (B) assay of MAPKK activity as previously described (16). These results are the average of three distinct experiments. VC, vector control.
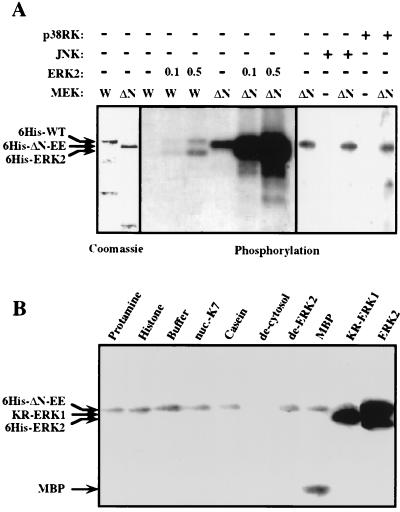
The distinct subcellular distribution observed with ΔN-EE-MEK could be due to severe changes that result in a new type of protein kinase with distinct activity, specificity, and, therefore, distinct localization. To test this hypothesis, two constructs (6His-ΔN-EE-MEK and 6His-WT-MEK) were prepared, expressed in bacteria, purified on a nickel column (Fig. 6A), and further characterized. As expected, both the recombinant proteins phosphorylated recombinant WT-ERK2 (Fig. 6A). However, the activity (≈0.1 nmol/min per mg) of 6His-WT-MEK was substantially lower than that of 6His-ΔN-EE-MEK (≈80 nmol/min per mg). In spite of the ΔN-EE-MEK’s elevated basal activity, its substrate specificity did not differ from that reported for purified, activated MEK (22). Only WT-ERK2 and KR-ERK1 (an inactive form of ERK1) were good substrates for ΔN-EE-MEK (Fig. 6B). MBP, as well as ΔN-EE-MEK, was also phosphorylated by the mutant kinase, but to a much smaller extent. The other nonrelated proteins and denatured ERK2 did not serve as substrates at all. Most important, although ERK1 and ERK2 were phosphorylated by ΔN-EE-MEK, other recombinant isoforms of MAPK (JNK and p38RK) were not (Fig. 6A) even at different pHs (data not shown). Therefore, the specificity of ΔN-EE-MEK does not seem to differ from that of WT-MEK.
Figure 6.
Substrate specificity of bacterially expressed 6His-ΔN-EE-MEK. 6His-ΔN-EE and 6His-WT-MEKs were expressed, purified, and subjected to Coomassie-blue staining (A Left). The two constructs (0.2 μg each) were used for in vitro phosphorylation (20 min, 30°C) of 0.1 μg, 0.5 μg, or no 6His-ERK2 (A Center). In a separate experiment, 6His-ΔN-EE-MEK (0.25 μg) was incubated under the same conditions with 6His-JNK1 (1 μg), 6His-p38RK (1 μg), or buffer alone. As control, the 6His-JNK1 and 6His-p38RK were incubated with buffer alone (A Right). W and ΔN indicate 6His-WT- and 6His-ΔN-EE-MEKs, respectively. (B) The proteins protamine, casein, histone (type III Sigma), nuclear K7 protein (gift from K. Bomsztyk, University of Washington), denatured extract (70°C, 10 min) of COS7 cells (de-cytosol, 10 μg), denatured (56°C, 10 min) ERK2 (de-ERK2, 0.5 μg), MBP, catalytically inactive ERK1 (6His-KR-ERK1, 0.5 μg), 6His-ERK2 (0.25 μg), or buffer A were subjected to in vitro phosphorylation by 6His-ΔN-EE-MEK (0.2 μg). The results in A and B were repeated three times.
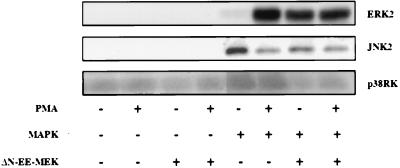
Although ΔN-EE-MEK does not activate kinases of other signaling cascades, a recent report suggests that ΔN-EE-MEK may activate the JNK cascade, probably via an autocrine loop (23). We tested this by cotransfecting into COS7 cells ΔN-EE-MEK together with HA-tagged isoforms of MAPK, such as HA-ERK2, HA-JNK2, or HA-p38RK. Two days posttransfection, the cells were harvested, the MAPKs immunoprecipitated with anti-HA antibodies, and the activity of each MAPK determined. Under these conditions, only ERK2 was activated, whereas the small basal activity of JNK2 and p38RK was slightly reduced (Fig. 7). Similar results were obtained with the in-gel kinase assay for ERK (24) and Jun phosphorylation (20) for JNK (data not shown). Therefore, our results indicate that no “cross-talk” occurs between the various cascades in these cells, which emphasizes the unique specificity of ΔN-EE-MEK. Overall, our results indicate that ΔN-EE-MEK and active WT-MEK do not differ in their specificity, and the differences in the localization of the mutants are probably due to the removal of the NES.
Figure 7.
ΔN-EE-MEK stimulates exclusively the ERK cascade in COS7 cells. Three HA-tagged MAPK isoforms (ERK2, JNK2, and p38RK) were cotransfected to COS7 cells with or without ΔN-EE-MEK followed by PMA stimulation (as in Fig. 5), immunoprecipitation using anti-HA antibodies, and kinase assay. The amount of phosphate incorporated into MBP in the stimulated ERK2 was 95-fold more than that in p38RK and 105-fold of that in c-Jun. This is a representative experiment that was repeated four times.
DISCUSSION
The identification of a special sequence that directs proteins out of the nucleus, termed NES (10, 11), drew our attention to a similar sequence located at residues 32–44 of MEK. In fact, when this sequence was deleted from the constitutively active, EE-MEK, the kinase changed its cytoplasmic localization to a nuclear one in both COS7 and HEK-293T cells (Figs. 2, 3, 4, 5). This subcellular distribution, which is also distinct from the normal cytosolic localization of WT-MEK, strongly indicates that the N-terminal sequence (amino acids 32–51) indeed functions as an NES. Similar findings have been recently reported by Fukuda et al. (9) that clearly demonstrate the involvement of the NES and particularly its leucine residues in the cellular localization of MEK in 3Y1 cells. These observations raise a question as to what could be the role of this NES sequence in the response of MEK to mitogenic stimulation. One possibility is that this sequence ensures the cytoplasmic localization of MEK and prevents its random diffusion into the nucleus. A more intriguing possibility is that MEK, like ERK, translocates to the nucleus upon activation, but that its apparent cytosolic distribution is a result of a rapid export from the nucleus that is driven by the NES. The small shift to the nucleus of WT- and EE-MEKs upon stimulation may have indicated a stimulation-induced translocation into the nucleus; however, the differences in staining between the basal and stimulated cells were too small to fully support such a conclusion. Moreover, the results obtained with ΔN-EE-MEK were also inconclusive, because the constitutive localization of this protein in the nucleus could have been due to the high enzymatic activity of ΔN-EE-MEK (Figs. 6 and 7), which might itself induce movement toward the nucleus. Therefore, the constitutive activity of MEK and its NES were separated to clarify whether nuclear translocation occurs in response to stimulation. ΔN-MEK possesses low, but significant, activity (16), and therefore, the ΔN-KA-EE-MEK, which completely lacks kinase activity (Fig. 5), and its NES region was used. Indeed, with the ΔN-KA-EE-MEK construct, most of the MEK was located in the cytosol of resting cells, but this shifted to the nucleus upon stimulation. Therefore, MEK appears to translocate to the nucleus upon stimulation in a mechanism that is distinct from its activation and is probably rapidly removed out of the nucleus by the NES. The role of this rapid movement in and out the nucleus is not clear, although the unique specificity of MEK toward ERK suggests that it involves an extra activation of this downstream kinase on their way into the nucleus. However, because no significant difference in ERK activity could be observed in nuclei from the different MEK mutant-expressing COS7 cells (not shown), it is possible that MEK phosphorylates other substrates in the nuclear envelope and in the cell periphery (possibly lamelopodia).
One of the key events in the transmission of intracellular signals is the translocation of protein kinases, such as protein kinase A (25), ERK, p90RSK (3), and others to the nucleus. This translocation enables the kinases to phosphorylate and activate transcription factors, and thus to regulate the expression of early genes (1). The mechanism by which these proteins translocate to the nucleus is not yet fully understood, because none of these kinases contains a NLS (26). The nuclear translocation of PKA was suggested to occur merely by diffusion (27). However, the rapid and quantitative translocation of ERK and p90RSK, as well as ΔN-KA-EE-MEK, into the nucleus makes it unlikely that simple diffusion is involved in this movement. It is therefore possible that the rapid movement of MEK into and out of the nucleus might be induced by an unknown mechanism that is common to many translocated kinases. However, it is also possible that MEK is translocated into the nucleus only when it is complexed with other signaling molecules or even by passive diffusion, and because it has no function there, it is rapidly exported out of this compartment.
In summary, our findings suggest that MEK translocates to the nucleus, and possibly also to the cell periphery, in response to mitogenic stimulation. However, MEK, unlike its downstream substrate ERK, seems to be rapidly exported out of the nucleus, due to a nuclear export signal sequence that is located in its N-terminal domain. The role of this rapid movement of MEK into and out of the nucleus is not clear, but it might involve an activation of ERK on its way into the nucleus.
Acknowledgments
We thank Dr. E. Schejter (Department of Molecular Genetics, The Weizmann Institute of Science) for his help in the confocal microscopy and Dr. B. Schick for helping in the preparation of the manuscript. This work was supported by grants from the Israeli Cancer Research Fund and the Israeli Cancer Association. R.S. is an incumbent of the Samuel and Isabela Friedman Career Development Chair.
ABBREVIATIONS
- MEK
mitogen-activated protein kinase kinase
- FCS
fetal calf serum
- MBP
myelin basic protein
- NES
nuclear export signal
- HA
hemagglutinin tag
- PMA
phorbol 12-myristate 13-acetate
- ERK
extracellular signal responsive kinase
References
- 1.Seger R, Krebs E G. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 2.Davis R J. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen R H, Sarnecki C, Blenis J. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenormand P, Sardet C, Pages G, L’Allemain G, Brunet A, Pouyssegur J. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marais R, Wynne J, Treisman R. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 7.Zheng C F, Guan K L. J Biol Chem. 1994;269:19947–19952. [PubMed] [Google Scholar]
- 8.Reszka A A, Seger R, Diltz C D, Krebs E G, Fischer E H. Proc Natl Acad Sci USA. 1995;92:8881–8885. doi: 10.1073/pnas.92.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda M, Gotoh I, Gotoh Y, Nishida E. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 11.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–73. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 12.Seger R, Seger D, Lozeman F J, Ahn N G, Graves L M, Campbell J S, Ericsson L, Harrylock M, Jensen A M, Krebs E G. J Biol Chem. 1992;267:25628–25636. [PubMed] [Google Scholar]
- 13.Seger R, Seger D, Reszka A A, Munar E S, Eldar-Finkelman H, Dobrowolska G, Jensen A M, Campbell J S, Fischer E H, Krebs E G. J Biol Chem. 1994;269:29876–29886. [PubMed] [Google Scholar]
- 14.Zheng C F, Guan K L. EMBO J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Wounde-Vande G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 17.Keown W A, Campbell C R, Kucherlapati R S. Methods Enzymol. 1990;185:527–537. doi: 10.1016/0076-6879(90)85043-n. [DOI] [PubMed] [Google Scholar]
- 18.Wigler M, Pellicer A, Silverstein S, Axel R. Cell. 1978;14:725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- 19.Crespo P, Xu N, Daniotti J L, Troppmair J, Rapp U R, Gutkind J S. J Biol Chem. 1994;269:21103–21109. [PubMed] [Google Scholar]
- 20.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 21.Dignam J D, Martin P L, Shastry B S, Roeder R G. Methods Enzymol. 1983;101:582–589. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 22.Seger R, Ahn N G, Posada J, Munar E S, Jensen A M, Cooper J A, Cobb M H, Krebs E G. J Biol Chem. 1992;267:14373–14381. [PubMed] [Google Scholar]
- 23.Franklin C C, Kraft A S. Oncogene. 1995;11:2365–2374. [PubMed] [Google Scholar]
- 24.Karunagaran D, Tzahar E, Beerli R R, Chen X, Graus P D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh D A, Brostrom C O, Brostrom M A, Chen L, Corbin J D, Reimann E, Soderling T R, Krebs E G. Adv Cyclic Nucleotide Res. 1972;1:33–45. [PubMed] [Google Scholar]
- 26.Akey C W. Semin Cell Biol. 1991;2:167–177. [PubMed] [Google Scholar]
- 27.Harootunian A T, Adams S R, Wen W, Meinkoth J L, Taylor S S, Tsien R Y. Mol Biol Cell. 1993;4:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]