Abstract
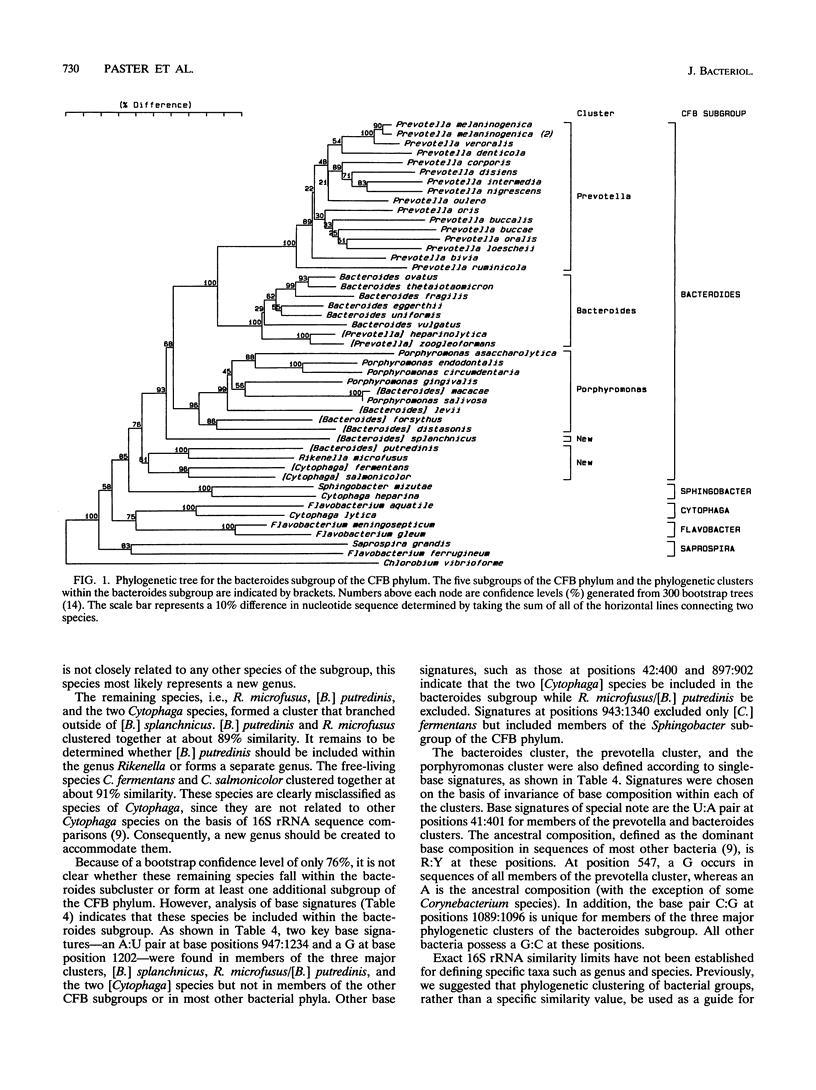
The phylogenetic structure of the bacteroides subgroup of the cytophaga-flavobacter-bacteroides (CFB) phylum was examined by 16S rRNA sequence comparative analysis. Approximately 95% of the 16S rRNA sequence was determined for 36 representative strains of species of Prevotella, Bacteroides, and Porphyromonas and related species by a modified Sanger sequencing method. A phylogenetic tree was constructed from a corrected distance matrix by the neighbor-joining method, and the reliability of tree branching was established by bootstrap analysis. The bacteroides subgroup was divided primarily into three major phylogenetic clusters which contained most of the species examined. The first cluster, termed the prevotella cluster, was composed of 16 species of Prevotella, including P. melaninogenica, P. intermedia, P. nigrescens, and the ruminal species P. ruminicola. Two oral species, P. zoogleoformans and P. heparinolytica, which had been recently placed in the genus Prevotella, did not fall within the prevotella cluster. These two species and six species of Bacteroides, including the type species B. fragilis, formed the second cluster, termed the bacteroides cluster. The third cluster, termed the porphyromonas cluster, was divided into two subclusters. The first contained Porphyromonas gingivalis, P. endodontalis, P. asaccharolytica, P. circumdentaria, P. salivosa, [Bacteroides] levii (the brackets around genus are used to indicate that the species does not belong to the genus by the sensu stricto definition), and [Bacteroides] macacae, and the second subcluster contained [Bacteroides] forsythus and [Bacteroides] distasonis. [Bacteroides] splanchnicus fell just outside the three major clusters but still belonged within the bacteroides subgroup. With few exceptions, the 16 S rRNA data were in overall agreement with previously proposed reclassifications of species of Bacteroides, Prevotella, and Porphyromonas. Suggestions are made to accommodate those species which do not fit previous reclassification schemes.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Booth S. J., Van Tassell R. L., Johnson J. L., Wilkins T. D. Bacteriophages of Bacteroides. Rev Infect Dis. 1979 Mar-Apr;1(2):325–336. doi: 10.1093/clinids/1.2.325. [DOI] [PubMed] [Google Scholar]
- Brondz I., Olsen I., Haapasalo M., Van Winkelhoff A. J. Multivariate analyses of fatty acid data from whole-cell methanolysates of Prevotella, Bacteroides and Porphyromonas spp. J Gen Microbiol. 1991 Jun;137(6):1445–1452. doi: 10.1099/00221287-137-6-1445. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Paster B. J., Olsen I., Fraser G. J. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992 Mar;174(6):2002–2013. doi: 10.1128/jb.174.6.2002-2013.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Wisotzkey J. D., Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992 Jan;42(1):166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- Gherna R., Woese C. R. A partial phylogenetic analysis of the "flavobacter-bacteroides" phylum: basis for taxonomic restructuring. Syst Appl Microbiol. 1992 Dec;15(4):513–521. doi: 10.1016/S0723-2020(11)80110-4. [DOI] [PubMed] [Google Scholar]
- Lambe D. W., Jr Determination of Bacteroides melaninogenicus serogroups by fluorescent antibody staining. Appl Microbiol. 1974 Oct;28(4):561–567. doi: 10.1128/am.28.4.561-567.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D. N., Bailey G. D., Collings S., Briscoe D. A. Description of Porphyromonas circumdentaria sp. nov. and reassignment of Bacteroides salivosus (Love, Johnson, Jones, and Calverley 1987) as Porphyromonas (Shah and Collins 1988) salivosa comb. nov. Int J Syst Bacteriol. 1992 Jul;42(3):434–438. doi: 10.1099/00207713-42-3-434. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993 Jan;7(1):113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., Dewhirst F. E., Weisburg W. G., Tordoff L. A., Fraser G. J., Hespell R. B., Stanton T. B., Zablen L., Mandelco L., Woese C. R. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991 Oct;173(19):6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza V., Tucker A. N., White D. C. Lipids of Bacteroides melaninogenicus. J Bacteriol. 1970 Jan;101(1):84–91. doi: 10.1128/jb.101.1.84-91.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shah H. N., Collins D. M. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990 Apr;40(2):205–208. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- Shah H. N., Gharbia S. E. Biochemical and chemical studies on strains designated Prevotella intermedia and proposal of a new pigmented species, Prevotella nigrescens sp. nov. Int J Syst Bacteriol. 1992 Oct;42(4):542–546. doi: 10.1099/00207713-42-4-542. [DOI] [PubMed] [Google Scholar]
- Shah H. N., Williams R. A. Dehydrogenase patterns in the taxonomy of Bacteroides. J Gen Microbiol. 1982 Dec;128(12):2955–2965. doi: 10.1099/00221287-128-12-2955. [DOI] [PubMed] [Google Scholar]
- Van den Eynde H., De Baere R., Shah H. N., Gharbia S. E., Fox G. E., Michalik J., Van de Peer Y., De Wachter R. 5S ribosomal ribonucleic acid sequences in Bacteroides and Fusobacterium: evolutionary relationships within these genera and among eubacteria in general. Int J Syst Bacteriol. 1989 Jan;39(1):78–84. doi: 10.1099/00207713-39-1-78. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Maloy S., Mandelco L., Raj H. D. Phylogenetic placement of the Spirosomaceae. Syst Appl Microbiol. 1990 Mar;13(1):19–23. doi: 10.1016/S0723-2020(11)80175-X. [DOI] [PubMed] [Google Scholar]
- van Steenbergen T. J., Vlaanderen C. A., de Graaff J. Deoxyribonucleic acid homologies among strains of Bacteroides melaninogenicus and related species. J Appl Bacteriol. 1982 Oct;53(2):269–276. doi: 10.1111/j.1365-2672.1982.tb04685.x. [DOI] [PubMed] [Google Scholar]


