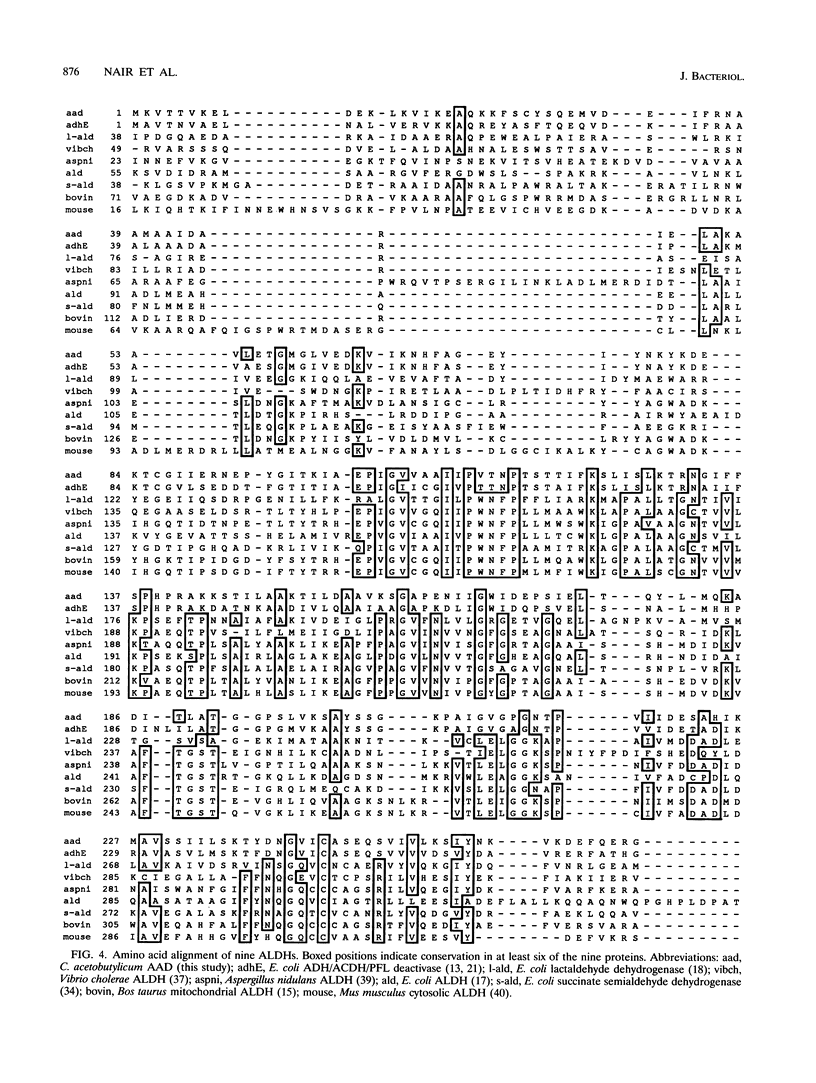
Abstract
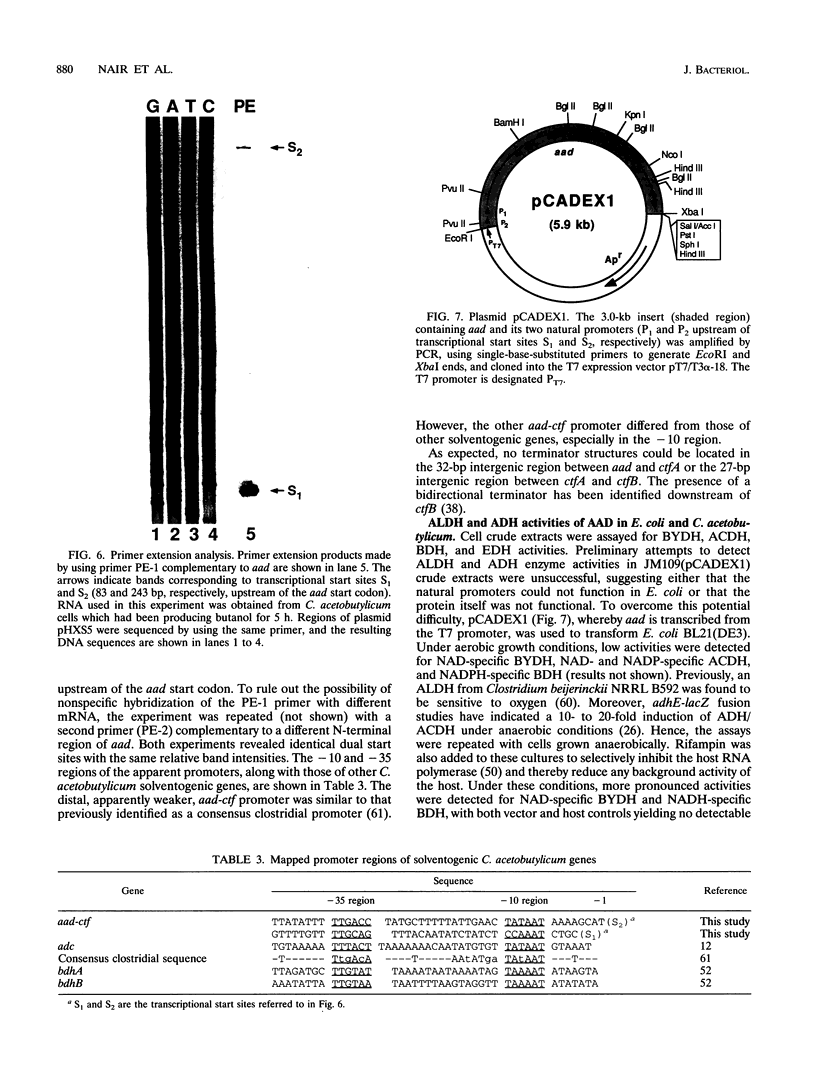
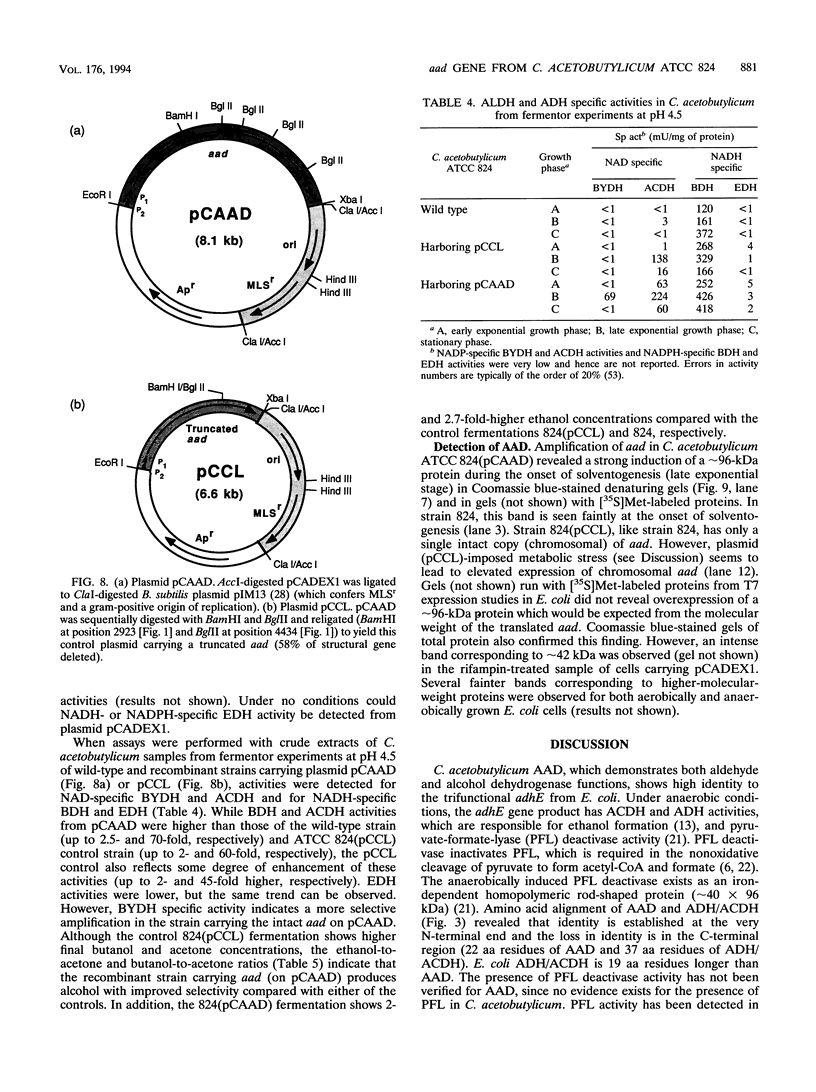
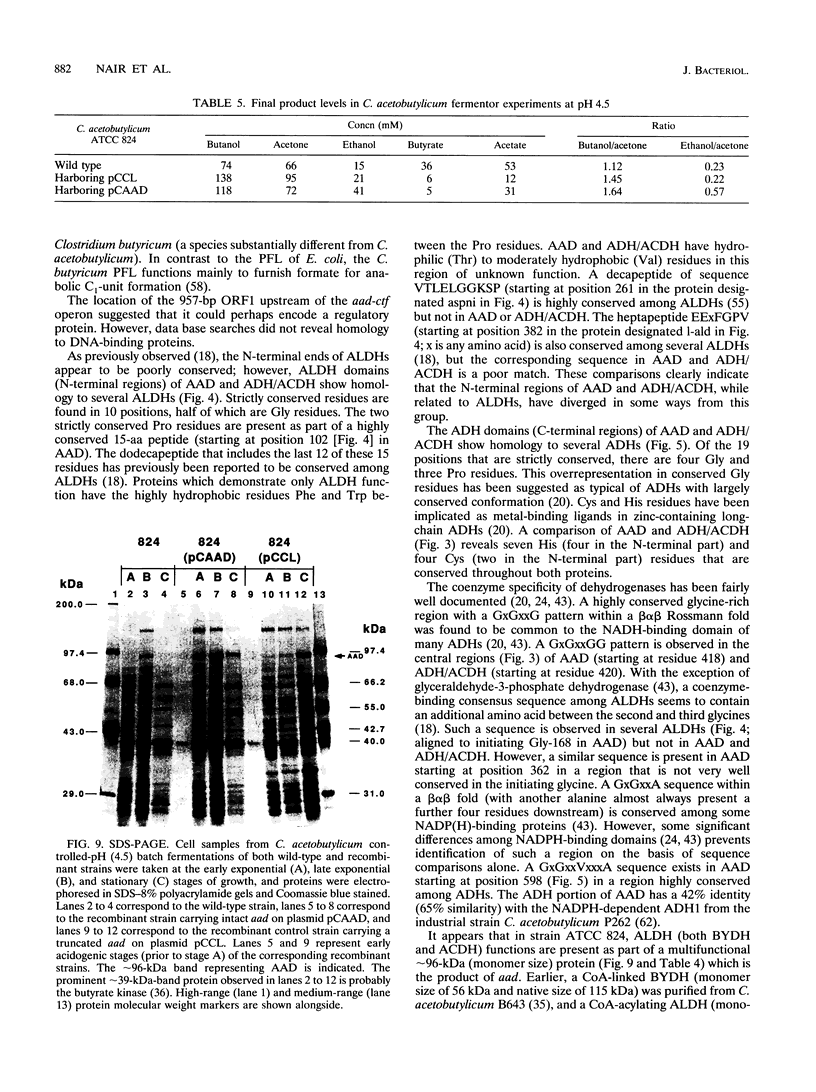
A gene (aad) coding for an aldehyde/alcohol dehydrogenase (AAD) was identified immediately upstream of the previously cloned ctfA (J. W. Cary, D. J. Petersen, E. T. Papoutsakis, and G. N. Bennett, Appl. Environ. Microbiol. 56:1576-1583, 1990) of Clostridium acetobutylicum ATCC 824 and sequenced. The 2,619-bp aad codes for a 96,517-Da protein. Primer extension analysis identified two transcriptional start sites 83 and 243 bp upstream of the aad start codon. The N-terminal section of AAD shows homology to aldehyde dehydrogenases of bacterial, fungal, mammalian, and plant origin, while the C-terminal section shows homology to alcohol dehydrogenases of bacterial (which includes three clostridial alcohol dehydrogenases) and yeast origin. AAD exhibits considerable amino acid homology (56% identity) over its entire sequence to the trifunctional protein encoded by adhE from Escherichia coli. Expression of aad from a plasmid in C. acetobutylicum showed that AAD, which appears as a approximately 96-kDa band in denaturing protein gels, provides elevated activities of NADH-dependent butanol dehydrogenase, NAD-dependent acetaldehyde dehydrogenase and butyraldehyde dehydrogenase, and a small increase in NADH-dependent ethanol dehydrogenase. A 957-bp open reading frame that could potentially encode a 36,704-Da protein was identified upstream of aad.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L. A., Adam L., Pelcher L. E., McHughen A., Hirji R., Selvaraj G. Characterization of an Escherichia coli gene encoding betaine aldehyde dehydrogenase (BADH): structural similarity to mammalian ALDHs and a plant BADH. Gene. 1991 Jul 15;103(1):45–52. doi: 10.1016/0378-1119(91)90389-s. [DOI] [PubMed] [Google Scholar]
- Cary J. W., Petersen D. J., Papoutsakis E. T., Bennett G. N. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase in Escherichia coli. Appl Environ Microbiol. 1990 Jun;56(6):1576–1583. doi: 10.1128/aem.56.6.1576-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Lu Z., Lin E. C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol. J Bacteriol. 1989 Nov;171(11):6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989 Sep;5(3):223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Clark S. W., Bennett G. N., Rudolph F. B. Isolation and Characterization of Mutants of Clostridium acetobutylicum ATCC 824 Deficient in Acetoacetyl-Coenzyme A:Acetate/Butyrate:Coenzyme A-Transferase (EC 2.8.3.9) and in Other Solvent Pathway Enzymes. Appl Environ Microbiol. 1989 Apr;55(4):970–976. doi: 10.1128/aem.55.4.970-976.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerischer U., Dürre P. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J Bacteriol. 1992 Jan;174(2):426–433. doi: 10.1128/jb.174.2.426-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlove P. E., Cunningham P. R., Parker J., Clark D. P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989 Dec 21;85(1):209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- Grupe H., Gottschalk G. Physiological Events in Clostridium acetobutylicum during the Shift from Acidogenesis to Solventogenesis in Continuous Culture and Presentation of a Model for Shift Induction. Appl Environ Microbiol. 1992 Dec;58(12):3896–3902. doi: 10.1128/aem.58.12.3896-3902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K. L., Weiner H. Sequence of the precursor of bovine liver mitochondrial aldehyde dehydrogenase as determined from its cDNA, its gene, and its functionality. Arch Biochem Biophys. 1990 Mar;277(2):351–360. doi: 10.1016/0003-9861(90)90590-u. [DOI] [PubMed] [Google Scholar]
- Guerrero F. D., Jones J. T., Mullet J. E. Turgor-responsive gene transcription and RNA levels increase rapidly when pea shoots are wilted. Sequence and expression of three inducible genes. Plant Mol Biol. 1990 Jul;15(1):11–26. doi: 10.1007/BF00017720. [DOI] [PubMed] [Google Scholar]
- Heim R., Strehler E. E. Cloning an Escherichia coli gene encoding a protein remarkably similar to mammalian aldehyde dehydrogenases. Gene. 1991 Mar 1;99(1):15–23. doi: 10.1016/0378-1119(91)90028-a. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Chen Y. M., Lin E. C., Aguilar J. Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J Bacteriol. 1991 Oct;173(19):6118–6123. doi: 10.1128/jb.173.19.6118-6123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Persson B., Jeffery J. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem. 1987 Sep 1;167(2):195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- Kessler D., Leibrecht I., Knappe J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991 Apr 9;281(1-2):59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- Knappe J., Sawers G. A radical-chemical route to acetyl-CoA: the anaerobically induced pyruvate formate-lyase system of Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):383–398. doi: 10.1111/j.1574-6968.1990.tb04108.x. [DOI] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Meulenberg C. H., Kingma J., Witholt B. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J Biol Chem. 1989 Apr 5;264(10):5442–5451. [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Blatterspiel R., Saleh M., Schiltz E., Schirmer R. H., Untucht-Grau R. Glutathione reductase from human erythrocytes. The sequences of the NADPH domain and of the interface domain. Eur J Biochem. 1982 Jan;121(2):259–267. doi: 10.1111/j.1432-1033.1982.tb05780.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leonardo M. R., Cunningham P. R., Clark D. P. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J Bacteriol. 1993 Feb;175(3):870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler I., Halvorson H. O. Two erythromycin-resistance plasmids of diverse origin and their effect on sporulation in Bacillus subtilis. J Gen Microbiol. 1980 Sep;120(1):259–263. doi: 10.1099/00221287-120-1-259. [DOI] [PubMed] [Google Scholar]
- Mermelstein L. D., Papoutsakis E. T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1993 Apr;59(4):1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein L. D., Welker N. E., Bennett G. N., Papoutsakis E. T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology (N Y) 1992 Feb;10(2):190–195. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- Narberhaus F., Giebeler K., Bahl H. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J Bacteriol. 1992 May;174(10):3290–3299. doi: 10.1128/jb.174.10.3290-3299.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palosaari N. R., Rogers P. Purification and properties of the inducible coenzyme A-linked butyraldehyde dehydrogenase from Clostridium acetobutylicum. J Bacteriol. 1988 Jul;170(7):2971–2976. doi: 10.1128/jb.170.7.2971-2976.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsakis E. T., Bennett G. N. Cloning, structure, and expression of acid and solvent pathway genes of Clostridium acetobutylicum. Biotechnology. 1993;25:157–199. [PubMed] [Google Scholar]
- Parsot C., Mekalanos J. J. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcriptional activator. J Bacteriol. 1991 May;173(9):2842–2851. doi: 10.1128/jb.173.9.2842-2851.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen D. J., Cary J. W., Vanderleyden J., Bennett G. N. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicum ATCC824. Gene. 1993 Jan 15;123(1):93–97. doi: 10.1016/0378-1119(93)90545-e. [DOI] [PubMed] [Google Scholar]
- Pickett M., Gwynne D. I., Buxton F. P., Elliott R., Davies R. W., Lockington R. A., Scazzocchio C., Sealy-Lewis H. M. Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene. 1987;51(2-3):217–226. doi: 10.1016/0378-1119(87)90310-6. [DOI] [PubMed] [Google Scholar]
- Rongnoparut P., Weaver S. Isolation and characterization of a cytosolic aldehyde dehydrogenase-encoding cDNA from mouse liver. Gene. 1991 May 30;101(2):261–265. doi: 10.1016/0378-1119(91)90421-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. T., Kaplan N. O. Purification, properties, and kinetic mechanism of coenzyme A-linked aldehyde dehydrogenase from Clostridium kluyveri. Arch Biochem Biophys. 1980 Sep;203(2):663–675. doi: 10.1016/0003-9861(80)90224-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Söhling B., Gottschalk G. Purification and characterization of a coenzyme-A-dependent succinate-semialdehyde dehydrogenase from Clostridium kluyveri. Eur J Biochem. 1993 Feb 15;212(1):121–127. doi: 10.1111/j.1432-1033.1993.tb17641.x. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter K. A., Bennett G. N., Papoutsakis E. T. Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J Bacteriol. 1992 Nov;174(22):7149–7158. doi: 10.1128/jb.174.22.7149-7158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. W., Rudolph F. B., Papoutsakis E. T. Purification and characterization of the NADH-dependent butanol dehydrogenase from Clostridium acetobutylicum (ATCC 824). Arch Biochem Biophys. 1989 Sep;273(2):309–318. doi: 10.1016/0003-9861(89)90489-x. [DOI] [PubMed] [Google Scholar]
- Weretilnyk E. A., Hanson A. D. Molecular cloning of a plant betaine-aldehyde dehydrogenase, an enzyme implicated in adaptation to salinity and drought. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2745–2749. doi: 10.1073/pnas.87.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenborn D. P., Rudolph F. B., Papoutsakis E. T. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl Environ Microbiol. 1988 Nov;54(11):2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Paquin C. E. Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol Gen Genet. 1987 Sep;209(2):374–381. doi: 10.1007/BF00329668. [DOI] [PubMed] [Google Scholar]
- Wood N. P., Jungermann K. Inactivation of the pyruvate formate lyase of Clostridium butyricum. FEBS Lett. 1972 Oct 15;27(1):49–52. doi: 10.1016/0014-5793(72)80407-1. [DOI] [PubMed] [Google Scholar]
- Wu L., Welker N. E. Cloning and characterization of a glutamine transport operon of Bacillus stearothermophilus NUB36: effect of temperature on regulation of transcription. J Bacteriol. 1991 Aug;173(15):4877–4888. doi: 10.1128/jb.173.15.4877-4888.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. T., Chen J. S. Coenzyme A-acylating aldehyde dehydrogenase from Clostridium beijerinckii NRRL B592. Appl Environ Microbiol. 1990 Sep;56(9):2591–2599. doi: 10.1128/aem.56.9.2591-2599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M., Minton N. P., Staudenbauer W. L. Recent advances in the genetics of the clostridia. FEMS Microbiol Rev. 1989 Dec;5(4):301–325. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]
- Youngleson J. S., Jones W. A., Jones D. T., Woods D. R. Molecular analysis and nucleotide sequence of the adh1 gene encoding an NADPH-dependent butanol dehydrogenase in the Gram-positive anaerobe Clostridium acetobutylicum. Gene. 1989 May 30;78(2):355–364. doi: 10.1016/0378-1119(89)90238-2. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries G. E., Arfman N., Terpstra P., Dijkhuizen L. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J Bacteriol. 1992 Aug;174(16):5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]