Abstract
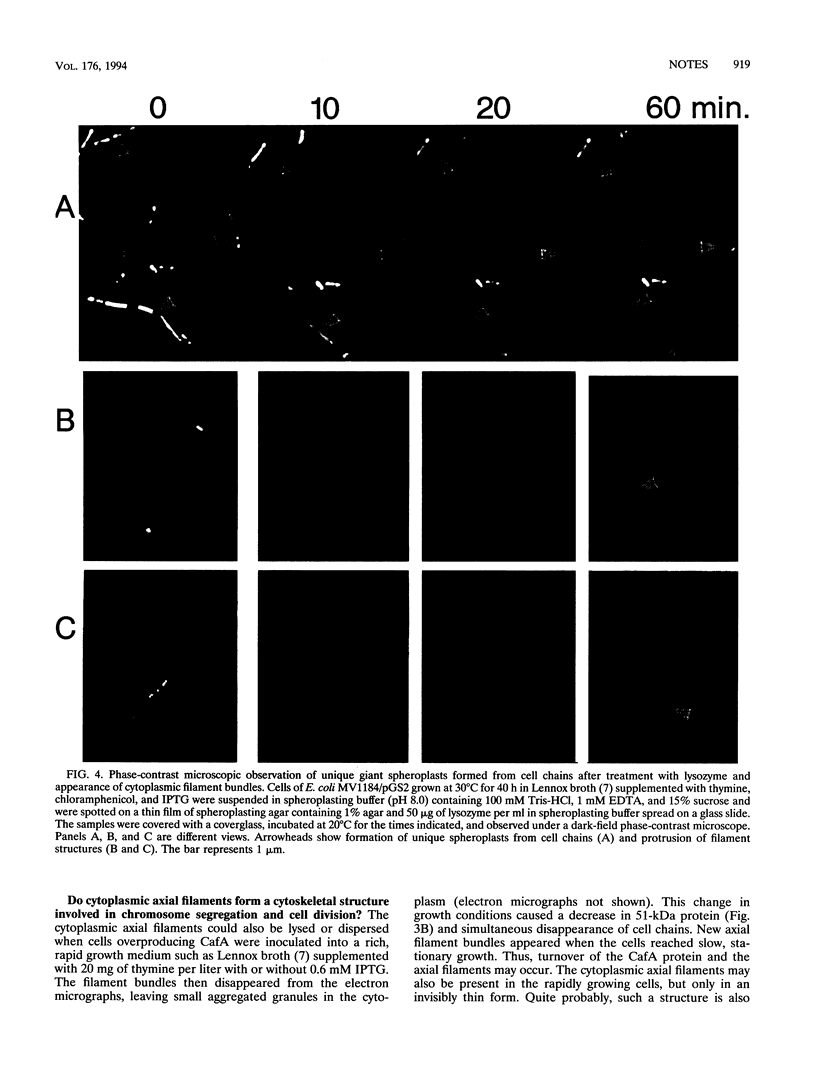
Overproduction of CafA caused formation of chained cells and minicells. The cafA gene is located downstream from the mre region at 71 min on the Escherichia coli chromosome map and was previously called orfF. A long axial structure running through the chained cells, consisting of bundles of filaments assembled in a long hexagonal pillar several micrometers long and about 0.1 to 0.2 micron in diameter, was visible in both phase-contrast micrographs of the lysozyme-treated cells and electron micrographs of ultrathin sections. The CafA protein displays 34% amino acid similarity with the N terminus of the Ams protein of E. coli, which cross-reacts with antibody to a nonmuscle myosin heavy chain.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casaregola S., Norris V., Goldberg M., Holland I. B. Identification of a 180 kD protein in Escherichia coli related to a yeast heavy-chain myosin. Mol Microbiol. 1990 Mar;4(3):505–511. doi: 10.1111/j.1365-2958.1990.tb00617.x. [DOI] [PubMed] [Google Scholar]
- Casarégola S., Jacq A., Laoudj D., McGurk G., Margarson S., Tempête M., Norris V., Holland I. B. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J Mol Biol. 1992 Nov 5;228(1):30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- Gayda R. C., Henk M. C., Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol. 1992 Aug;174(16):5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Ogura T., Niki H., Ichinose C., Mori H. Positioning of replicated chromosomes in Escherichia coli. J Bacteriol. 1990 Jan;172(1):31–39. doi: 10.1128/jb.172.1.31-39.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobot J. A., Carlemalm E., Villiger W., Kellenberger E. Periplasmic gel: new concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. J Bacteriol. 1984 Oct;160(1):143–152. doi: 10.1128/jb.160.1.143-152.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- McDowall K. J., Hernandez R. G., Lin-Chao S., Cohen S. N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993 Jul;175(13):4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H., Jaffé A., Imamura R., Ogura T., Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991 Jan;10(1):183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W., Larson T. J. Physical locations of genes in the rne (ams)-rpmF-plsX-fab region of the Escherichia coli K-12 chromosome. J Bacteriol. 1992 Dec;174(23):7873–7874. doi: 10.1128/jb.174.23.7873-7874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L. I., DeBoer P., Cook W. R. Localization of septation sites. Res Microbiol. 1990 Jan;141(1):57–63. doi: 10.1016/0923-2508(90)90098-b. [DOI] [PubMed] [Google Scholar]
- Spurr A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969 Jan;26(1):31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wachi M., Doi M., Okada Y., Matsuhashi M. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol. 1989 Dec;171(12):6511–6516. doi: 10.1128/jb.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Doi M., Tamaki S., Park W., Nakajima-Iijima S., Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987 Nov;169(11):4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Doi M., Ueda T., Ueki M., Tsuritani K., Nagai K., Matsuhashi M. Sequence of the downstream flanking region of the shape-determining genes mreBCD of Escherichia coli. Gene. 1991 Sep 30;106(1):135–136. doi: 10.1016/0378-1119(91)90578-y. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]







