Abstract
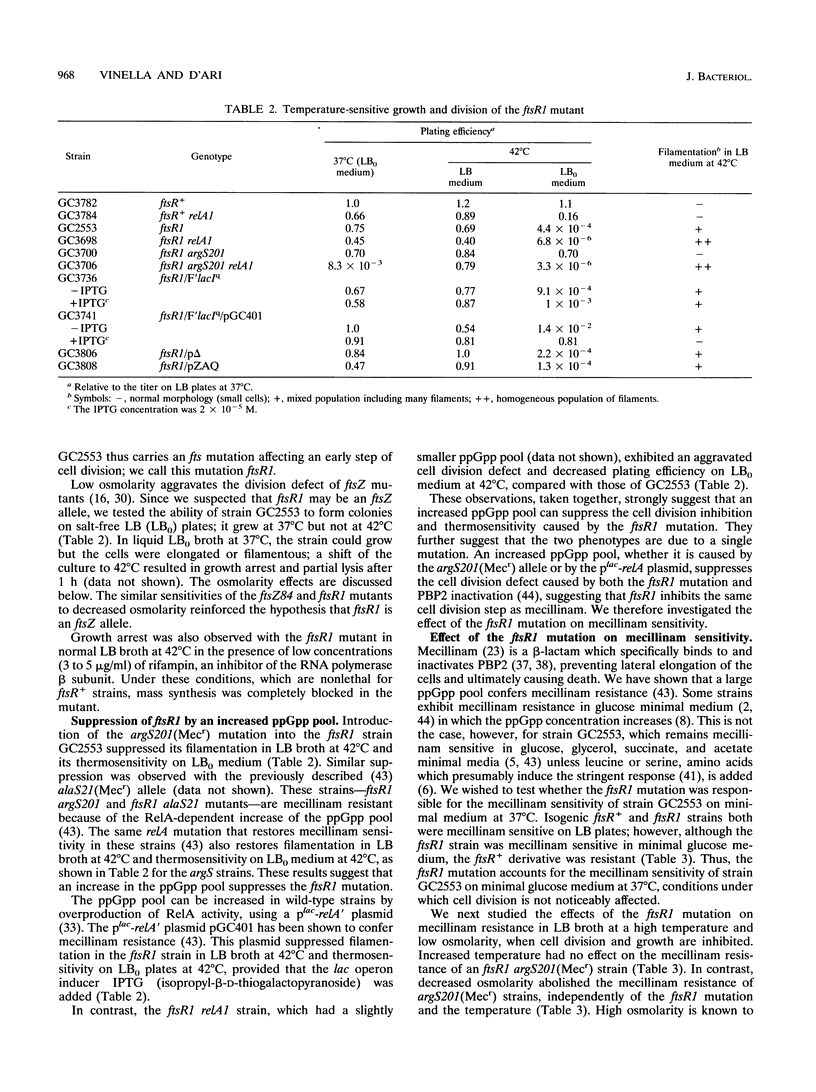
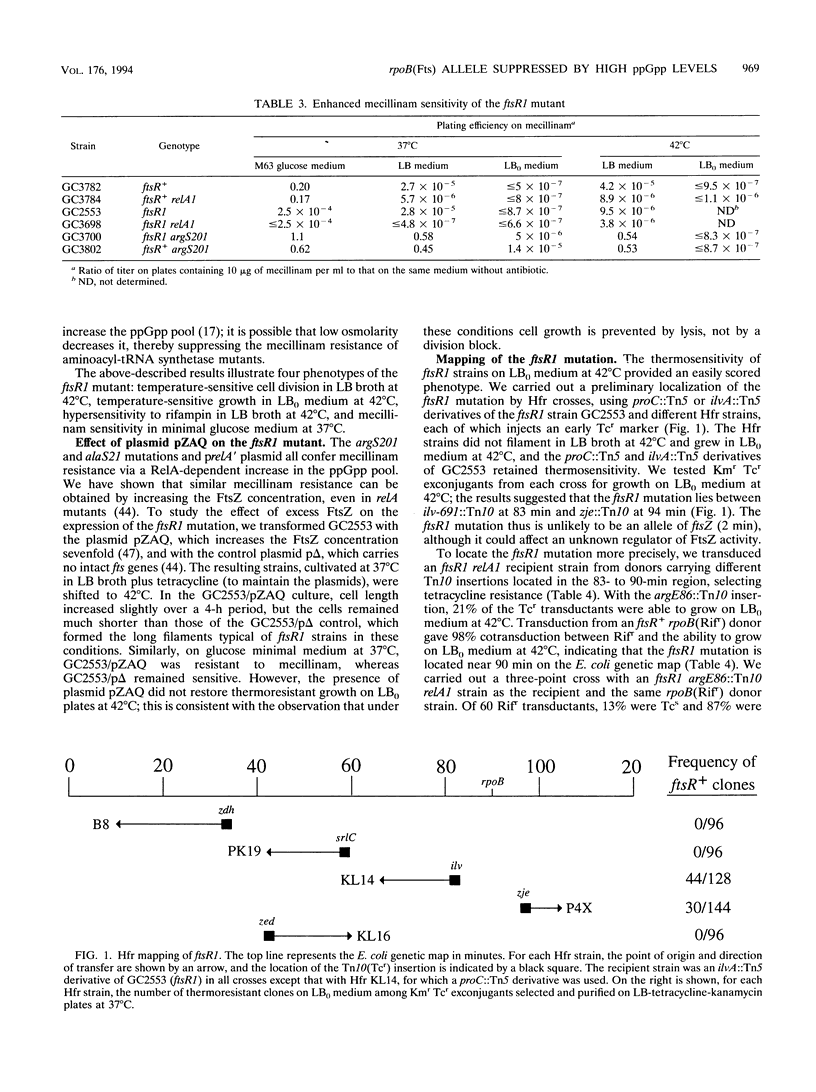
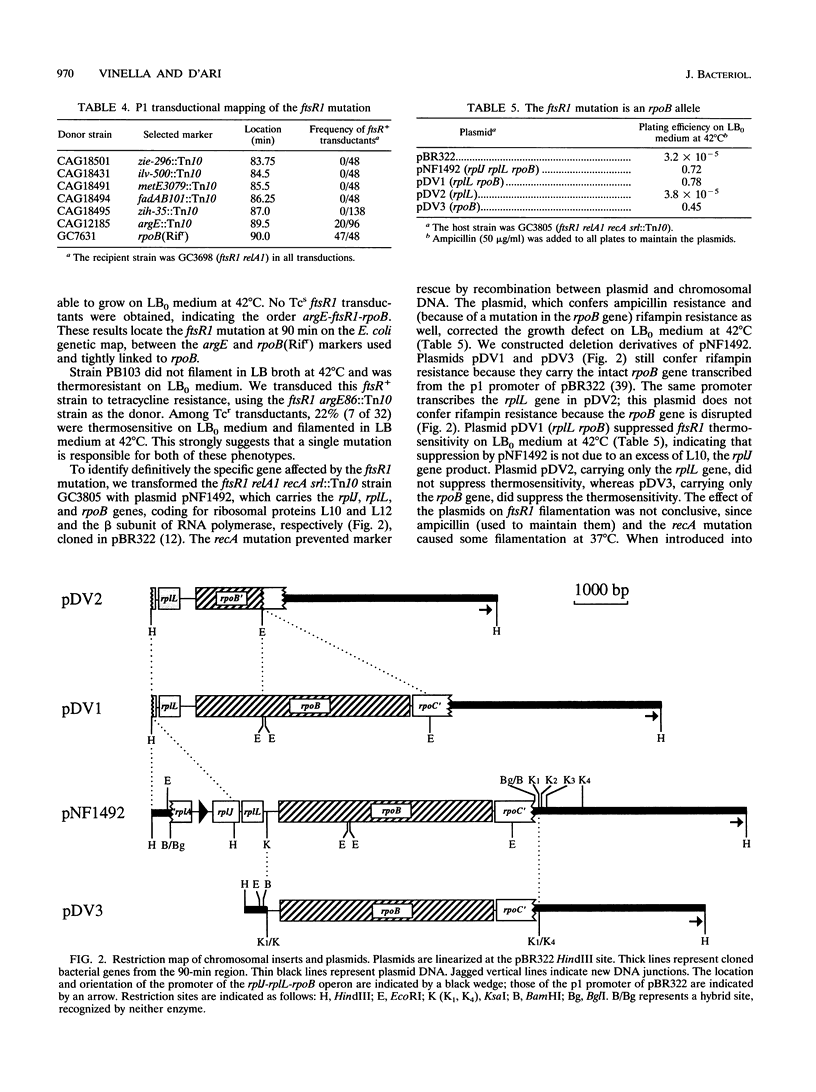
The Escherichia coli strain known as GC2553, FB8, UTH1038, or K12S (Luria), considered an F- lambda- wild-type strain, is shown here to carry a cryptic mutation, ftsR1, causing nonlethal filamentation during exponential growth in Luria-Bertani (LB) broth at 42 degrees C and the inability to grow in salt-free LB broth at 42 degrees C. The ftsR1 mutation is completely suppressed in genetic backgrounds which increase RelA-dependent synthesis of the nucleotide ppGpp, i.e., argS201 (Mecr) and alaS21 (Mecr) mutations, affecting aminoacyl-tRNA synthetases, or the presence of a plac-relA' plasmid. These backgrounds also confer resistance in LB broth to the beta-lactam mecillinam, an antibiotic which specifically inhibits penicillin-binding protein 2 and, in wild-type cells, causes an indirect block in cell division. Furthermore, the ftsR1 mutant (but not an isogenic ftsR+ strain) is sensitive to mecillinam in minimal glucose medium at 37 degrees C. Since the division block caused by mecillinam can be overcome by overproduction of the cell division protein FtsZ, we tested the effect of plasmid pZAQ (carrying the ftsZ, ftsA, and ftsQ genes) on the ftsR1 mutant; it suppressed the filamentation in LB broth and the mecillinam sensitivity on minimal glucose medium at 37 degrees C but not the growth defect in salt-free LB broth at 42 degrees C. Genetic analysis indicated that the full phenotype of the ftsR1 mutant is due to a single mutation in the rpoB gene (90 min), coding for the beta subunit of RNA polymerase; we call this allele rpoB369(Fts). We propose that the rpoB369(Fts) mutation alters the specificity of the polymerase and that the mutant enzyme can recover normal activity in the presence of high salt concentrations or via interaction with the nucleotide ppGpp.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baracchini E., Glass R., Bremer H. Studies in vivo on Escherichia coli RNA polymerase mutants altered in the stringent response. Mol Gen Genet. 1988 Aug;213(2-3):379–387. doi: 10.1007/BF00339606. [DOI] [PubMed] [Google Scholar]
- Barbour A. G., Mayer L. W., Spratt B. G. Mecillinam resistance in Escherichia coli: dissociation of growth inhibition and morphologic change. J Infect Dis. 1981 Jan;143(1):114–121. doi: 10.1093/infdis/143.1.114. [DOI] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991 Nov 14;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993 Feb;175(4):1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloc P., Jaffé A., D'Ari R. The Escherichia coli lov gene product connects peptidoglycan synthesis, ribosomes and growth rate. EMBO J. 1989 Jan;8(1):317–323. doi: 10.1002/j.1460-2075.1989.tb03379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloc P., Vinella D., D'Ari R. Leucine and serine induce mecillinam resistance in Escherichia coli. Mol Gen Genet. 1992 Nov;235(2-3):242–246. doi: 10.1007/BF00279366. [DOI] [PubMed] [Google Scholar]
- Bruni C. B., Colantuoni V., Sbordone L., Cortese R., Blasi F. Biochemical and regulatory properties of Escherichia coli K-12 hisT mutants. J Bacteriol. 1977 Apr;130(1):4–10. doi: 10.1128/jb.130.1.4-10.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. X., Bouquin N., Norris V., Casarégola S., Séror S. J., Holland I. B. A single base change in the acceptor stem of tRNA(3Leu) confers resistance upon Escherichia coli to the calmodulin inhibitor, 48/80. EMBO J. 1991 Oct;10(10):3113–3122. doi: 10.1002/j.1460-2075.1991.tb07865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil N. P., Bendiak D., Collins J., Friesen J. D. Expression of Escherichia coli ribosomal protein and RNA polymerase genes cloned on plasmids. Mol Gen Genet. 1979 May 23;173(1):39–50. doi: 10.1007/BF00267689. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Jones S. T., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VII. RNA polymerase is a target for ppGpp. Mol Gen Genet. 1986 May;203(2):265–268. doi: 10.1007/BF00333964. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Jones S. T., Nomura T., Ishihama A. Hierarchy of the strength of Escherichia coli stringent control signals. Mol Gen Genet. 1987 Nov;210(1):1–4. doi: 10.1007/BF00337750. [DOI] [PubMed] [Google Scholar]
- Goldschmidt E. P., Cater M. S., Matney T. S., Butler M. A., Greene A. Genetic analysis of the histidine operon in Escherichia coli K12. Genetics. 1970 Oct;66(2):219–229. doi: 10.1093/genetics/66.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Halpern E., Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981 Oct;148(1):265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman R. B., Yamazaki H. MSI accumulation induced by sodium chloride. Biochemistry. 1972 Feb 15;11(4):615–618. doi: 10.1021/bi00754a023. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989 Mar;171(3):1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Huisman O., D'Ari R., Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc G., Sirard C., Drapeau G. R. The Escherichia coli cell division mutation ftsM1 is in serU. J Bacteriol. 1989 Apr;171(4):2090–2095. doi: 10.1128/jb.171.4.2090-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R., Ryals J., Bremer H. Physiological characterization of Escherichia coli rpoB mutants with abnormal control of ribosome synthesis. J Bacteriol. 1983 Sep;155(3):1162–1170. doi: 10.1128/jb.155.3.1162-1170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund F., Tybring L. 6 -amidinopenicillanic acids--a new group of antibiotics. Nat New Biol. 1972 Apr 5;236(66):135–137. doi: 10.1038/newbio236135a0. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Regulation of cell division in E. coli. Trends Genet. 1990 Jan;6(1):22–25. doi: 10.1016/0168-9525(90)90045-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y., Yamagata H. Sucrose-dependent spectinomycin-resistant mutants of Escherichia coli. J Bacteriol. 1976 Jan;125(1):142–148. doi: 10.1128/jb.125.1.142-148.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Dai K., Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Relaxed mutants of Escherichia coli RNA polymerase. FEBS Lett. 1983 Mar 21;153(2):307–310. doi: 10.1016/0014-5793(83)80630-9. [DOI] [PubMed] [Google Scholar]
- Ogura T., Bouloc P., Niki H., D'Ari R., Hiraga S., Jaffé A. Penicillin-binding protein 2 is essential in wild-type Escherichia coli but not in lov or cya mutants. J Bacteriol. 1989 Jun;171(6):3025–3030. doi: 10.1128/jb.171.6.3025-3030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix P., Drapeau G. R. Cell division control in Escherichia coli K-12: some properties of the ftsZ84 mutation and suppression of this mutation by the product of a newly identified gene. J Bacteriol. 1988 Sep;170(9):4338–4342. doi: 10.1128/jb.170.9.4338-4342.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D., Park J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992 Sep 17;359(6392):251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- SPOTTS C. R., STANIER R. Y. Mechanism of streptomycin action on bacteria: a unitary hypothesis. Nature. 1961 Nov 18;192:633–637. doi: 10.1038/192633a0. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Metzger S., Aizenman E., Roza S., Cashel M., Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991 Feb 25;266(6):3760–3767. [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Pardee A. B. Penicillin-binding proteins and cell shape in E. coli. Nature. 1975 Apr 10;254(5500):516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- Spratt B. G. The mechanism of action of mecillinam. J Antimicrob Chemother. 1977 Jul;3 (Suppl B):13–19. doi: 10.1093/jac/3.suppl_b.13. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K., Bremer H. Toxic effects of high levels of ppGpp in Escherichia coli are relieved by rpoB mutations. J Biol Chem. 1992 Feb 5;267(4):2337–2344. [PubMed] [Google Scholar]
- Uzan M., Danchin A. Correlation between the serine sensitivity and the derepressibility of the ilv genes in Escherichia coli relA- mutants. Mol Gen Genet. 1978 Sep 20;165(1):21–30. doi: 10.1007/BF00270372. [DOI] [PubMed] [Google Scholar]
- Van der Meide P. H., Borman T. H., Van Kimmenade A. M., Van de Putte P., Bosch L. Elongation factor Tu isolated from Escherichia coli mutants altered in TufA and tufB. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3922–3926. doi: 10.1073/pnas.77.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., D'Ari R., Jaffé A., Bouloc P. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992 Apr;11(4):1493–1501. doi: 10.1002/j.1460-2075.1992.tb05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Joseleau-Petit D., Thévenet D., Bouloc P., D'Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J Bacteriol. 1993 Oct;175(20):6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. R., Kovarik A., Allen J. S., Gustafson R. A. Regulation of bacterial cell division: temperature-sensitive mutants of Escherichia coli that are defective in septum formation. J Bacteriol. 1975 Aug;123(2):693–703. doi: 10.1128/jb.123.2.693-703.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992 Sep 17;359(6392):254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]



