Abstract
One of the main uncertainties in risk estimation for environmental radon exposure using lung cancer data from underground miners is the extrapolation from high- to low-dose exposure where multiple traversal is extremely rare. The biological effects of a single α particle are currently unknown. Using the recently available microbeam source at the Radiological Research Accelerator Facility at Columbia University, we examined the frequencies and molecular spectrum of S1− mutants induced in human–hamster hybrid (AL) cells by either a single or an exact number of α particles. Exponentially growing cells were stained briefly with a nontoxic concentration of Hoechst dye for image analysis, and the location of individual cells was computer-monitored. The nucleus of each cell was irradiated with either 1, 2, 4, or 8 α particles at a linear energy transfer of 90 keV/μm consistent with the energy spectrum of domestic radon exposure. Although single-particle traversal was only slightly cytotoxic to AL cells (survival fraction ≈ 0.82), it was highly mutagenic, and the induced mutant fraction averaged 110 mutants per 105 survivors. In addition, both toxicity and mutant induction were dose-dependent. Multiplex PCR analysis of mutant DNA showed that the proportion of mutants with multilocus deletions increased with the number of particle traversals. These data provide direct evidence that a single α particle traversing a nucleus will have a high probability of resulting in a mutation and highlight the need for radiation protection at low doses.
Accurate risk assessment of human exposure to ionizing radiations traditionally has been compromised, in that reliable data are available only for relatively high doses, so that extrapolations must be made down to the relevant, low-dose region of interest in radiation protection. However, this approach in risk assessment is often complicated by concurrent exposure to other chemical and physical environmental contaminants. Data indicate that exposure of the lung to α-emitting radon progeny is the largest component of background radiation received by the general public in the United States (1). Epidemiological studies have shown that uranium miners exposed to high levels of radon progeny have the largest incidence of radiation-induced lung cancers of any exposed population (2, 3). However, studies designed to identify a link between lung cancer and the low levels of radon commonly found in the home have been inconclusive because of confounding factors. The recent estimate by the Environmental Protection Agency of 21,600 deaths per year (confidence limits between 7,000 and 30,000) illustrates the uncertainties inherent in environmental risk assessment using epidemiological data (see ref. 4 for review).
Radon, a secondary decay product of uranium-238, is a colorless, odorless gas that decays with a half-life of 3.82 days into a series of solid, short-lived radionucleotides, including polonium-218 and polonium-214 that emit α particles during decay. Radon is ubiquitous in indoor environments, including homes and schools, and, in general, at concentrations hundreds of fold lower than in underground mines.
To have a better quantitative assessment of lung cancer risk associated with residential radon exposure, it is essential to have a better database for low-dose exposure. It has been estimated that 96% of the target bronchial cells of an average uranium miner will be traversed by more than one α particle each year. In contrast, only 1 in 107 bronchial cells will be hit by multiple particles from an average household exposure (4). The biological effects of a single α-particle traversal are unknown. Several relevant questions arise: Is a single traversal by these high linear energy transfer (LET) particles lethal to a cell? If not, will the surviving cells have a higher propensity to undergo chromosomal aberrations, mutations, and neoplastic transformation than nonirradiated cells? How does the number of particle traversals affect the kinds of mutations induced? The availability of a microbeam irradiation facility at the Radiological Research Accelerator Facility at Columbia University, where individual cells can be irradiated with either a single or an exact number of α particles, provides a unique opportunity to address these questions.
Since individual cells are irradiated one at a time so as to limit the number of cells available for analysis, a sensitive mutagenic assay system is essential to give meaningful data. The AL cells developed by Waldren and Puck (5) fulfill this requirement. These cells contain a standard set of hamster chromosomes, but only one human chromosome (chromosome 11), which carries specific cell-surface antigenic markers. By the use of appropriate antibodies, mutations in the human chromosome can be quantified. Because only a small segment of this human chromosome (11p15.5) is needed for viability of the hybrid cell, this mutation system is particularly sensitive to agents such as ionizing radiations and asbestos fibers that induce multilocus deletions (6, 7). The AL surface antigens (S1, S2) are effective genetic markers, because their presence or absence can be easily measured, and their distribution on opposite arms of chromosome 11 permits identification of lesions involving the long, short, or both chromosome arms. In the present studies, we have determined the dose response with regard to toxicity, mutant induction, and the kinds of mutations at the S1 locus found in cells whose nuclei were exposed to either a single or an exact number of α particles. Our data provide the first demonstration that a single α particle hit in the nucleus, which kills only 20% of the cells, is indeed mutagenic.
MATERIALS AND METHODS
Cell Culture.
The AL hybrid cells that contain a standard set of Chinese hamster ovary-K1 chromosomes and a single copy of human chromosome 11 were used. Chromosome 11 encodes cells surface markers that render AL cells sensitive to killing by specific monoclonal antibodies in the presence of complement. Rabbit serum complement was from HPR (Denver, PA). Antibody specific to the S1 antigen was produced from hybridoma culture as described (5, 8, 9). Cells were maintained in Ham F-12 medium supplemented with 8% heat-inactivated fetal bovine serum, 25 μg/ml gentamycin, and 2× normal glycine (2 × 10−4M) at 37°C in a humidified 5% CO2 incubator, and passaged as described (6, 7).
Irradiation Procedure.
The layout and irradiation procedure using the microbeam facility at the Radiological Research Accelerator Facility have been described (10). Approximately 500 exponentially growing AL cells were inoculated into each of a series of microbeam dishes constructed by drilling a ¼-inch hole in the center of 60-mm-diameter non-tissue-culture dishes. A 3.8-μm-thick polypropylene film was epoxied over the bottom of the hole, creating a miniwell that was then coated with Cel-Tak to enhance cell attachment. The DNA of attached cells was stained with a 50 nM solution of Hoechst 33342 dye for 30 min, and the location of individual nuclei was determined by optical imaging of the fluorescent staining pattern at 366 nm. The image analysis system then located the centroid of each nucleus, which were irradiated one at a time with an exact number of α particles. On average, it took 2 sec to locate and irradiate a cell so that up to 10,000 cells could be irradiated per day. We used 15,000–20,000 irradiated cells per group per experiment in the present study. The overall spatial precision of the beam, including positioning and beam spread, is about ±4 μm. Because the average cross-sectional area of the nucleus of live, attached AL cells was determined to be 108 μm2, we estimated by Monte Carlo modeling of the collimators that the particle beam would hit the targeted nucleus 98.4% of the time. Due to the lag time of shutter closure, about 4 per 1,000 nuclei would have received one extra α particle. After every cell on a plate had been irradiated, the dish was removed from the stage, and the cells were trypsinized and replated to measure both survival and mutation as described (6, 11, 12). The percent recovery of irradiated cells from the polypropylene dishes was >98% as determined by cell count of representative dishes.
Dose Response for Cytotoxicity.
Irradiated and control cells recovered from each miniwell were trypsinized and replated into 100-mm-diameter Petri dishes for colony formation. Cultures were incubated for 7–12 days, at which time they were fixed with formaldehyde and stained with Giemsa. The number of colonies was counted to determine the surviving fraction as described (11, 12).
Quantification of Mutations at the S1 Locus.
Irradiated and control cultures from each miniwell were replated into 25-cm2-area tissue-culture flasks and cultured for 7 days. This expression period is needed to permit surviving cells to recover from the temporary growth lag caused by irradiation and to multiply such that the progeny of the mutated cells no longer express lethal amounts of the S1 surface antigen. To determine mutant fractions, 5 × 104 cells were plated into each of six 60-mm dishes in a total of 2.5 ml of growth medium as described (6, 7, 11). The cultures were incubated for 2 h to allow for cell attachment, after which 0.2% S1 antiserum and 1.5% freshly thawed complement (vol/vol) were added to each dish. After overnight incubation, the medium in each dish was changed to remove the antiserum and complement. The cultures were further incubated for 7 to 8 days, at which time they were fixed, stained, and the number of S1− mutants scored. Controls included identical sets of dishes containing antiserum alone, complement alone, or neither agent. The cultures derived from each well were tested for mutant yield for 2 consecutive weeks to ensure full expression of the mutations. Mutant fractions were calculated as the number of surviving colonies divided by the total number of cells plated after correction for any nonspecific killing due to complement alone.
Analysis of Mutant Spectrum by Multiplex PCR.
S1− mutants were isolated by cloning and expanded in cultures as described (6, 7, 11). We isolated no more than two well-separated colonies per culture dish to ensure their clonal origin. A minimum of 25 mutants from each irradiated group and over 50 spontaneous mutants were analyzed. DNA was extracted by a simple salting-out procedure as described (11, 13).
Five DNA markers on chromosome 11 (Wilms tumor, parathyroid hormone, catalase, RAS, and apolipoprotein A-1) were chosen for multiplex PCR analysis because of their map positions relative to the MIC1 gene, which codes for the S1 antigen (14), and the availability of PCR primers for the coding regions of these genes (15–17). PCR amplifications were performed for 30 cycles using a DNA Thermal Cycler 480 (Perkin–Elmer Cetus) in 20-μl reaction mixtures containing 1 μg of the EcoRI-digested DNA sample in 1× Stoffel fragment buffer, 0.2 mM dNTP, 3 mM MgCl2, 0.2 mM each primer, and 2 units of Stoffel fragment enzyme. Each PCR cycle consisted of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. After the last cycle, the samples were incubated at 72°C for an additional 20 min, electrophoresed on 2% agarose gels, and stained with ethidium bromide.
RESULTS
Irradiation of Cell Nuclei with the Microbeam.
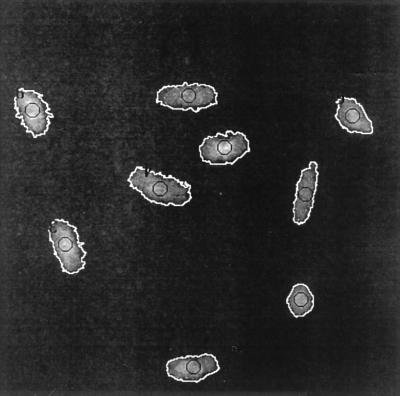
Fig. 1 shows the fluorescent image of a representative population of AL cells as seen by the image analysis system under a 40× objective lens. Each nucleus was outlined by the image analysis program, and the precise center of each nucleus was located and placed over the exit aperture of the beam. Selected numbers of α particles then were automatically delivered within the 5 μm diameter of the beam line. These areas are shown by the small circles in the center of each nucleus (Fig. 1). Because the particle detector was positioned behind the monolayer of cells, every α particle registered would have traversed the nucleus. Given the track length of ≈40 μm for the 5.5-MV α particles used here when they entered attached mammalian cells, and the thickness of the AL cell nucleus measured to be less than 4 μm, it was unlikely that any α particle would be stopped within the nucleus. The pixel position of irradiated nuclei was recorded to prevent multiple exposure of the same cell. It should be noted that the 50 nM dose of Hoechst dye used in the imaging step had been determined to be nontoxic and nonmutagenic under the conditions used in these studies§ and verified here.
Figure 1.
Fluorescent imaging of AL cells stained with Hoechst dye viewed by the image analysis system under a 40× objective lens. The nucleus of each cell is outlined in white, and the circles indicate the area where the particle is delivered.
Lethality of a Single-Particle Traversal.
There is considerable interest in the carcinogenic effects of low doses of high LET radiations, such as α particles. It has been estimated for attached mammalian cells, for example, that the mean number of α-particle traversals required for cell killing ranges from 2 to 6 (18). Direct measurement of the lethality of a single α particle was, until recently, not possible. Fig. 2 shows the dose–response clonogenic survival of AL cells irradiated with defined numbers of α traversals through the nucleus. The curve was best fitted by a linear quadratic model with α = 0.285 ± 0.01 and β = 0, yielding a mean lethal dose of ≈3.7 particles. It is clear from these data that most of the cells (≈80%) survived to form colonies after exposure of their nuclei to a single particle. In fact, more than 10% of the cells survived after nuclear traversal by eight particles.
Figure 2.
Survival of AL cells irradiated in the nucleus with either a single or an exact number of α particles. Data were pooled from 3–4 independent experiments. Bar represents ± SEM.
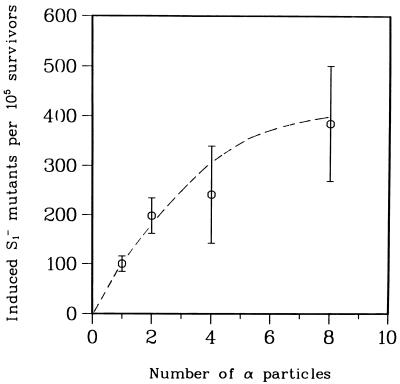
Mutagenicity of a Single α Particle.
The relatively high mutagenic sensitivity of the AL cell system made it possible to assess the mutagenic potential of a single α particle from relatively few irradiated cells. Mutation data were analyzed using the least-square method with the following parameters: y = 108x − 7.7x2 where y was the number of induced mutants and x represented the number of particle traversals. Fig. 3 shows the number of induced mutant (background subtracted) per 105 clonogenic survivors at the S1 locus in AL cells irradiated with either a single or an exact number of α particles. The fraction of preexisting S1− mutants in the AL cell population used in these experiments averaged 45 per 105 survivors. The induced mutant yield by a single α particle was 2-fold of this background level and increased to 8-fold for eight particles. The dose–response curve yielded an initial slope of ≈100 mutants/105 cells per particle.
Figure 3.
Induced mutants per 105 survivors at the S1 locus in AL cells irradiated with an exact number of α-particle traversals at 90 keV/μm. Induced mutant yield = total mutant yield minus background incidence. The background mutant fraction in AL cells used in these experiments averaged 45 per 105 survivors. Data were pooled from three experiments, and the curves fitted using the least-square method. (Bars represent ± SEM.)
Analysis of Mutant Spectrum.
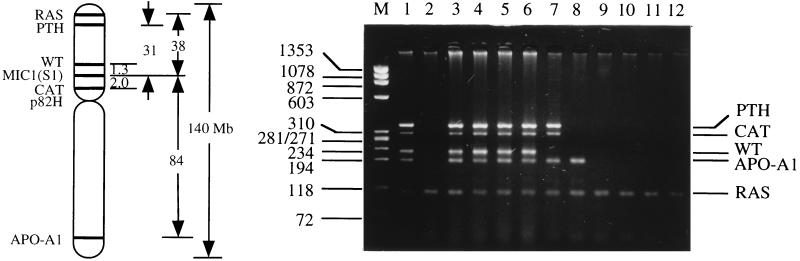
The S1 surface antigenic marker is encoded by the MIC1 gene mapped to chromosome 11p13. With the mapping of over 200 genes on both the short and long arm of chromosome 11, together with the availability of primer sequences for some of these genes, it is relatively easy to determine the spectrum of S1− mutants induced. A total of 167 mutants, including 57 spontaneous ones, were analyzed. Fig. 4 shows a representative gel of PCR products using DNA from S1− mutants as template and primers synthesized for specific regions of marker genes located on either the long arm (Apo-A1) or short arm (CAT, WT, PTH, and RAS) of the human chromosome 11. The presence or absence of the corresponding PCR products indicates that the particular segments of DNA containing these genes are present or missing, respectively. Fig. 5 shows the cumulative deletion maps of these S1− mutants. Previous studies have shown that a small segment of the human chromosome 11 near the RAS gene is required for survival of the S1− mutants (11, 19). The obligate presence of this region identified here by the RAS probe in all the mutants provides a convenient internal PCR control. Fig. 4 shows that the majority of spontaneous S1− mutants (50 of 57 or 88%) had retained all of the markers analyzed. Likewise, the majority of mutants induced by a single α particle resulted from mutations involving the loss of S1 marker only (24 of 32 or 75%), whereas the remaining 25% of the mutants had lost at least one additional marker. In contrast, the proportion of mutants suffering loss of additional chromosomal markers increased with increasing number of particle traversals such that 19 of 24 (79%) of the mutants induced by eight particles had lost all four markers examined, which spanned both arms of the human chromosome 11. These mutants were further characterized by Southern blotting using the centromeric probe p82H (20). Approximately 50% of the mutants had lost the centromere, indicating a loss of the entire human chromosome 11 except for the 11p15.5 fragment that had translocated to a hamster chromosome (data not shown).
Figure 4.
(Right) Gel electrophoresis of multiplex PCR products using DNA from S1− mutants as templates and primers for parathyroid hormone (PTH), Wilms tumor (WT), catalase (CAT), apolipoprotein A1 (APO-A1), and RAS. HaeIII-digested φX174 DNA provided the size markers (lane M). Lane 1, wild-type AL cells with all of the markers present. Lane 2, a positive control showing the loss of all the markers examined except RAS. Lanes 3 and 4, spontaneous mutants showing no marker loss. Lanes 5 and 6, mutants induced by a single α particle where none of these markers was lost. Lanes 7–12, mutants induced by eight α particles showing mostly deletions of various sizes. (Left) The relative location of the marker genes on human chromosome 11 used in the multiplex PCR and their relative distance from the M1C1 gene.
Figure 5.
Cumulative deletion spectra of S1− mutants either of spontaneous origin or from cells exposed to either a single or an exact number of α-particle traversals through the nucleus. Each line depicts the spectrum from a single, independent mutant. The absence or presence of marker genes among the mutants was determined by multiplex PCR. Blank spaces depict missing markers.
DISCUSSION
It is of societal importance to provide realistic risk estimate for the carcinogenic effects of domestic radon exposure, currently estimated at 15,000 lung cancer deaths per year. This number is based largely on extrapolation from the high-dose exposure data for underground miners where the majority of the target bronchial epithelial cells received multiple α-particle traversals (4). However, environmental radon exposure levels are such that multiple traversals are extremely rare, so that the effects of a single α-particle hit are the most relevant to environmental risk analysis (21). Our data provide a direct measurement of the genotoxicity of a single α particle.
The question of whether traversal of a single α particle through the nucleus is lethal has been debated for more than three decades. Earlier studies by Barendsen (22) suggested that traversal of the nucleus by a single α particle would be lethal. Moreover, studies based on measurement of induced DNA double-strand breaks in C3H10T½ cells indicated that virtually 100% of the cells traversed by a single α particle would be killed by direct action (23). On the other hand, microdosimetric studies based on particle track structure suggested that the probability of an α-particle traversal resulting in lethal damage was only 17% in rodent fibroblasts, i.e. it takes six hits to kill a cell (24). Our direct measurement of a single particle survival is consistent with an estimate of a low probability of cell inactivation: only 20% of the irradiated cells were killed. It is amazing that roughly 10% of cells irradiated with eight α particles were still viable enough to form colonies even though they carried a much higher mutagenic potential.
Using the nuclear crosssectional area of 108 μm2 measured for AL cells, we calculated that a dose of ≈12 cGy of 90 keV/μm α particles from track segment irradiation where attached cells are exposed to a broad beam of monoenergenetic particles would be required to deliver an average of one particle traversal per nucleus based on random, Poisson distribution. At this dose, about one-third of the nuclei would not be hit, another third would sustain one α-particle hit, and the remaining third would receive multiple hits. The dose response for survival of AL cells irradiated with an exact number of α particles was not significantly different from recent data obtained using average particle traversals (11, 12). These results suggest that, at least for cell lethality, the Poisson estimation gives a fairly accurate projection of the biological effects of either a single or an exact number of α particles.
The numbers and kinds of mutants induced by α particles at several gene loci, including thymidine kinase and hypoxanthine-guanine phosphoribosyltransferase, have been reviewed recently (25, 26). High LET radiation, such as α particles, induced more mutant per mean lethal dose (Do) than low LET radiation such as x- and γ-rays (≈280 S1− mutants/Do for 90 keV/μm α particles versus 150 mutants/Do for γ-rays; refs. 11, 12). The number of induced mutants is both dose- and LET-dependent. Our present data provide the first demonstration that a single α particle induces mutations in mammalian cells. Using the highly sensitive AL assay system, we were able to show that a single-particle traversal induced a mutant fraction 2 times greater than the background value. This mutant yield was comparable to the frequency induced by an equivalent mean of one particle traversal based on a Poisson distribution (11). Our results are consistent with those of Nelson et al. (27) who demonstrated a linear dose response with regard to the induction of micronuclei among Chinese hamster ovary cells irradiated with up to five α particles using a 3.2-MV microbeam. However, at a dose of eight particle traversals per nucleus, where we found the induced frequency was 8 times background level, the incidence was significantly higher than the yield obtained with a mean of eight particles as determined by the Poisson distribution (data not shown). It is possible that many cells in the latter group may received either very few particle traversals and subsequently fewer mutations, or many more than eight particles that are lethal to the cells. Thus, it is likely that this difference is due to distortion of the cell population at the time of the assay in the track segment experiment (Poisson distributed) because of differences in radiation-induced division delay. The cells that received a small number of traversals would be expected to expand more rapidly during the expression period than those that received a large number of particle hits. Thus, there is a closed correlation between the effects of exactly one and a mean of one particle, the single cell irradiation allows a more accurate extrapolation from high to low doses.
While the majority of radiation-induced mutants showed deletions of varying sizes (11, 28, 29), there is recent evidence to indicate that the percentage of multilocus deletions is dose- and LET- dependent as well (11, 12). As shown in Fig. 4, the majority of spontaneous S1 mutants (88%) have lost only the S1 marker, presumably as a result of either a point mutation or a small deletion involving the MIC1 gene. These findings are consistent with our previous studies when only a limited number of marker genes were used (7, 11). However, due to the distance of the two nearest marker gene (WT and CAT) from the M1C1 gene, mutants that are classified as S1− only have the potential of losing up to a 3.3-Mb region of the human chromosome 11. As the number of particle traversals increase, the relative proportion of mutants losing only the S1 marker decreased from 75% among mutants induced by a single particle to 4% among those induced by eight α particles. Because the mutant fraction induced by a single α particle was two times higher than the spontaneous background, it is likely that one out of three mutants analyzed could be of spontaneous origin. Nevertheless, the mutant spectrum from the single-particle group was not significantly different from that of spontaneous. This data suggest that most mutants induced by a single particle harbored deletions that were smaller than those traversed by multiple particles. These results further confirmed our previous finding that the relative proportion of mutants with large gene/chromosomal deletions is dose-dependent (11, 12).
Acknowledgments
We thank Dr. You-ping Huang of our center for assistance with data analysis. This work was supported by National Cancer Institute Grants CA 49092, CA 36447, and CA 56392, National Institutes of Health Research Resource Center Grant RR 11623, National Research Service Award CA64039, National Aeronautics and Space Administration Contract NAF 9501-0232, and the Waldren/Vannais Laboratory Fund. T.K.H. is a joint faculty member of the Department of Environmental Health Sciences, Columbia University School of Public Health, and C.W. is a member of the Cancer Center, University of Colorado School of Medicine.
ABBREVIATION
- LET
linear energy transfer
Footnotes
Hei, T. K., Wu, L.-J., Liu, S.-X., Vannais, D., Waldren, C. & Randers-Pehrson, G. Proceedings of the Annual Radiation Research Society Meeting, April 14–17, 1996, Chicago, IL.
References
- 1.National Council on Radiation Protection and Measurements. National Council on Radiation Protection and Measurements Report No. 79. Washington, DC: National Academy of Sciences; 1984. [Google Scholar]
- 2.Lundin F E, Lloyd J W, Smith E M, Archer V E, Holaday D A. Health Phys. 1969;16:571–578. doi: 10.1097/00004032-196905000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Whittemore A S, McMillan A. J Natl Cancer Inst. 1983;71:489–493. [PubMed] [Google Scholar]
- 4.National Research Council Committee on Health Risks of Exposure to Radon (BEIR VI) Health Effects of Exposure to Radon: Time for Reassessment. Washington, DC: National Academy of Sciences; 1994. [Google Scholar]
- 5.Waldren C, Jones C, Puck T T. Proc Natl Acad Sci USA. 1979;76:1358–1362. doi: 10.1073/pnas.76.3.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hei T K, Waldren C A, Hall E J. Radiat Res. 1988;115:281–291. [PubMed] [Google Scholar]
- 7.Hei T K, Piao C Q, Zhu L X, He Z Y, Vannais D, Waldren C A. Cancer Res. 1992;52:6305–6309. [PubMed] [Google Scholar]
- 8.Puck T T, Wuchier P, Jones C, Kao F T. Proc Natl Acad Sci USA. 1971;68:3102–3106. doi: 10.1073/pnas.68.12.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldren C, Correll L, Sognier M A, Puck T T. Proc Natl Acad Sci USA. 1986;83:4839–4843. doi: 10.1073/pnas.83.13.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randers-Pherson G. In: Annual Report of the Center for Radiological Research. Kliauga P, editor. 1995. pp. 35–39. [Google Scholar]
- 11.Zhu L X, Waldren C A, Vannais D, Hei T K. Radiat Res. 1996;145:251–259. [PubMed] [Google Scholar]
- 12.Hei T K, Zhu L X, Vannais D, Waldren C A. Adv Space Res. 1994;14:355–361. doi: 10.1016/0273-1177(94)90488-x. [DOI] [PubMed] [Google Scholar]
- 13.Miller S A, Dykes D D, Polesky H F. Nucleic Acid Res. 1988;16:1215–1219. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones C, Moore E E, Lehman D W. Proc Natl Acad Sci USA. 1979;76:6491–6495. doi: 10.1073/pnas.76.12.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier J, Bruening W, Kashtan C E, Mauer S M, Manivel J C, Striegel E, Houghton D C, Junien C, Habib R, Fouser L, Fine R N, Silverman B L, Haber D A, Housman D E. Cell. 1991;67:437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 16.Vasicek T J, McDevitt B E, Freeman M W, Fennick B J, Hendy O N, Potts J T, Jr, Rich A, Kronenberg H M. Proc Natl Acad Sci USA. 1983;80:2127–2131. doi: 10.1073/pnas.80.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karathanasis S K, Zannis V I, Breslow J L. Proc Natl Acad Sci USA. 1983;80:6147–6151. doi: 10.1073/pnas.80.20.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raju M R, Eisen Y, Carpenter S, Inkret W C. Radiat Res. 1991;128:204–209. [PubMed] [Google Scholar]
- 19.McGuinness S M, Shibuya S M, Ueno A M, Vannais D, Waldren C A. Radiat Res. 1995;142:247–255. [PubMed] [Google Scholar]
- 20.Waye J S, Creeper L A, Willard H F. Chromosoma. 1987;11:182–188. doi: 10.1007/BF00330349. [DOI] [PubMed] [Google Scholar]
- 21.Lubin J H, Liang Z, Hrubec Z, Pershagen G, Schoenberg J B, Blot W J, Klotz J B, Xu J, Boice J D. Cancer Causes Control. 1994;5:114–128. doi: 10.1007/BF01830257. [DOI] [PubMed] [Google Scholar]
- 22.Barendsen G W. Int J Radiat Biol. 1964;8:453–466. doi: 10.1080/09553006414550561. [DOI] [PubMed] [Google Scholar]
- 23.Watt D E. Radiat Prot Dosim. 1989;27:73–84. [Google Scholar]
- 24.Roberts C J, Goodhead D T. Int J Radiat Biol. 1987;52:871–882. doi: 10.1080/09553008714552461. [DOI] [PubMed] [Google Scholar]
- 25.Evans H H. Radiat Res. 1994;137:131–144. [PubMed] [Google Scholar]
- 26.Little J B. Radiat Res. 1994;140:299–311. [PubMed] [Google Scholar]
- 27.Nelson J M, Brooke A L, Metting N F, Khan M A, Buschbom R L, Duncan A, Miick R, Braby L A. Radiat Res. 1996;145:568–574. [PubMed] [Google Scholar]
- 28.Thacker J. Mutat Res. 1986;160:267–275. doi: 10.1016/0027-5107(86)90137-5. [DOI] [PubMed] [Google Scholar]
- 29.Jostes R F, Fleck E W, Morgan T L, Stiegler G L, Cross F T. Radiat Res. 1994;137:371–379. [PubMed] [Google Scholar]