Abstract
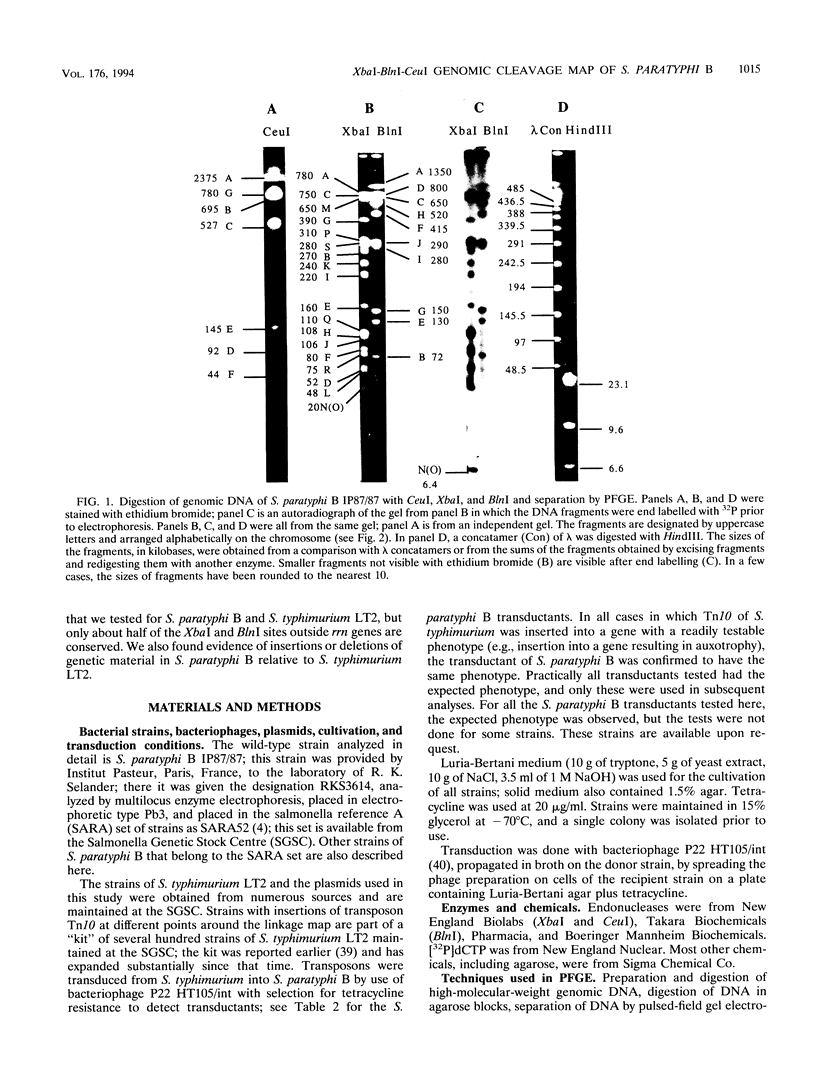
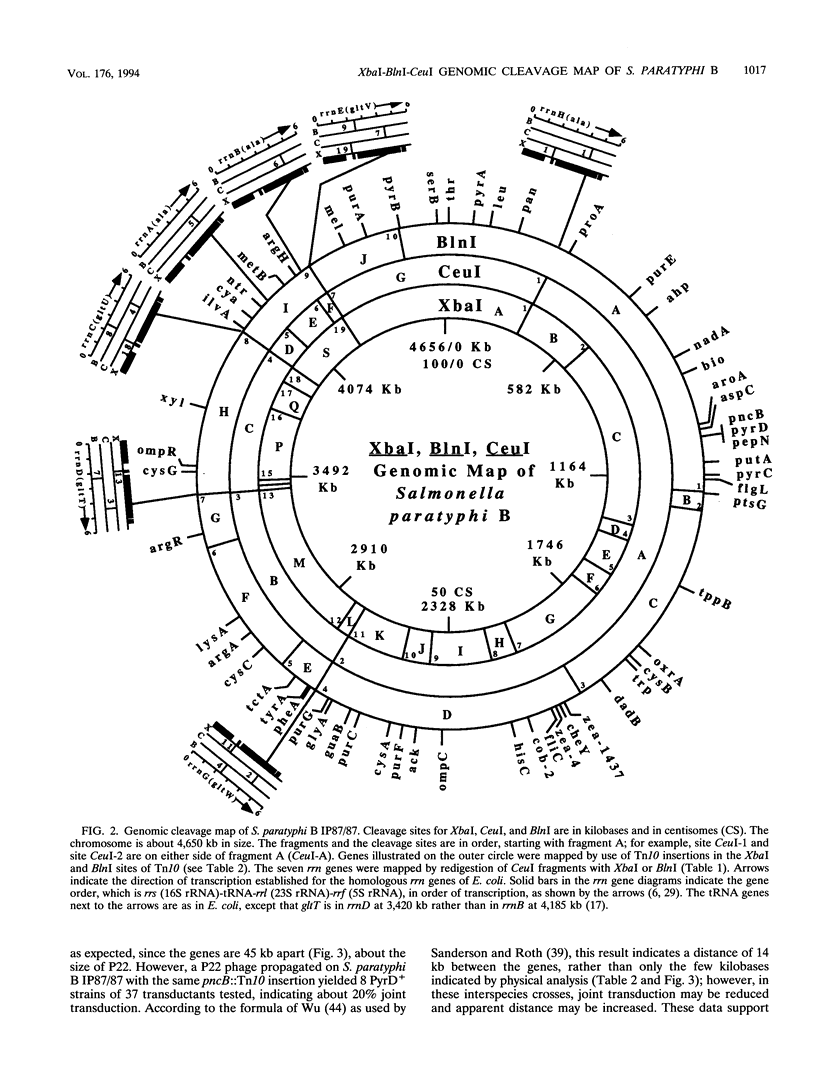
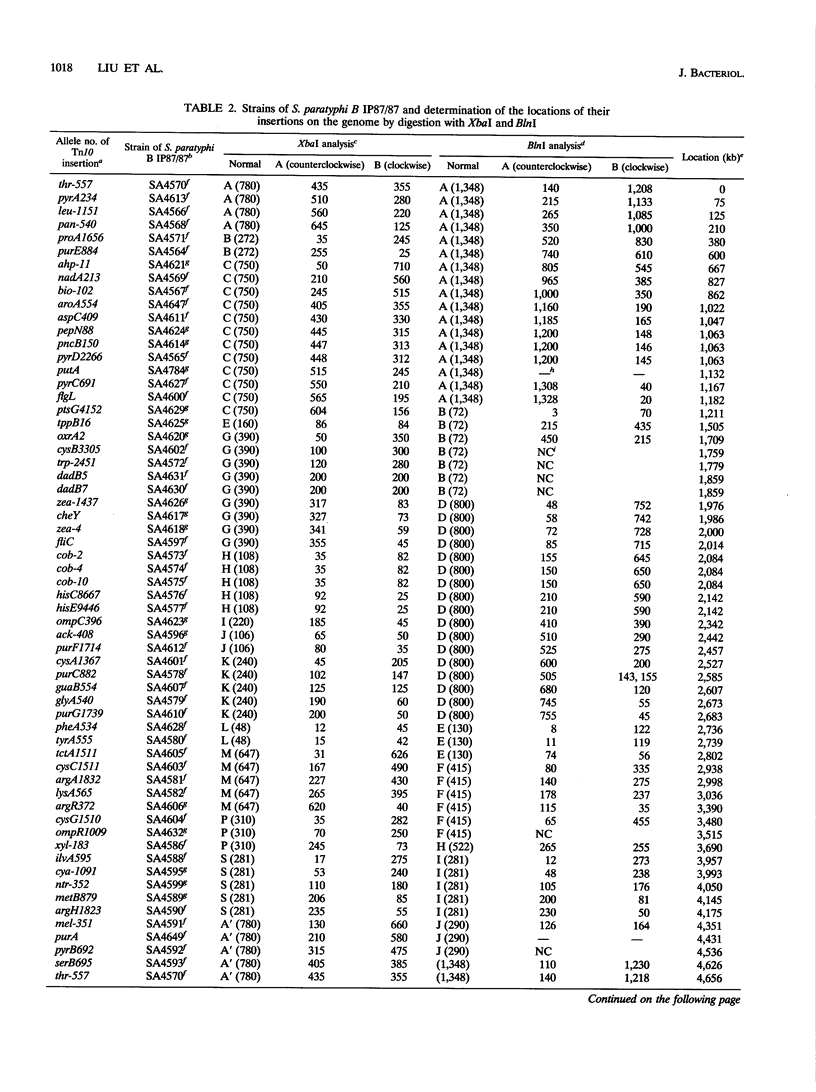
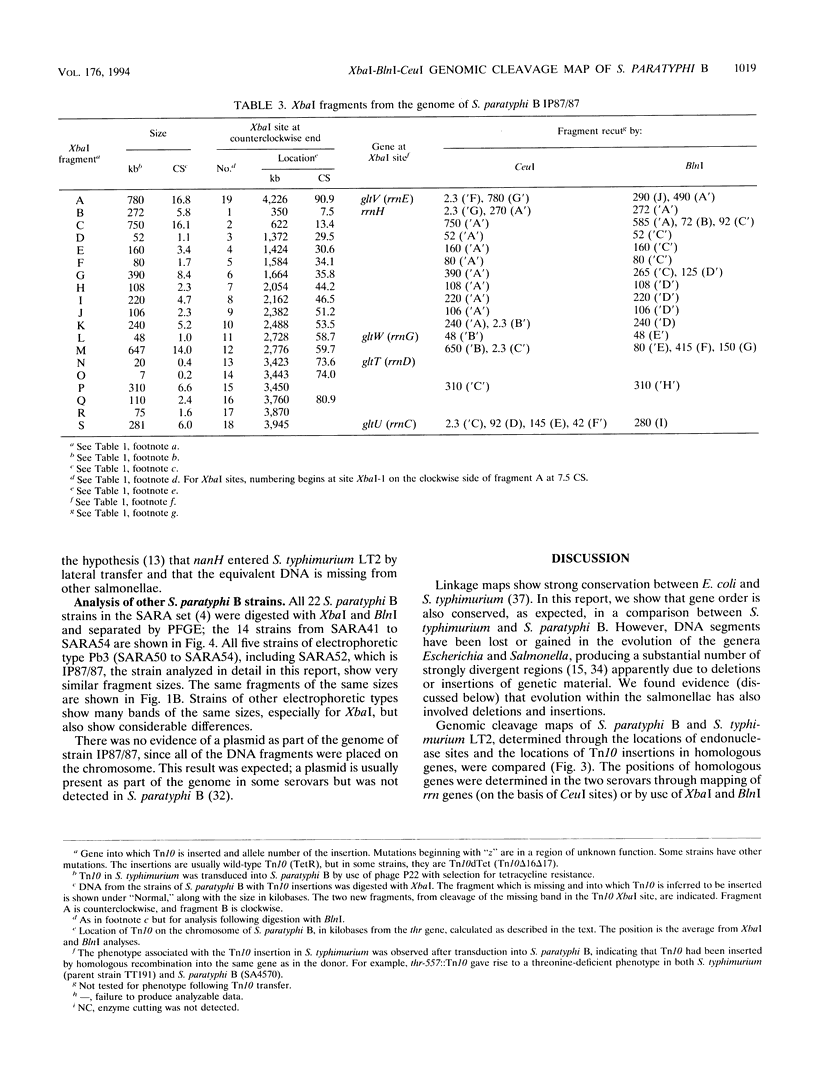
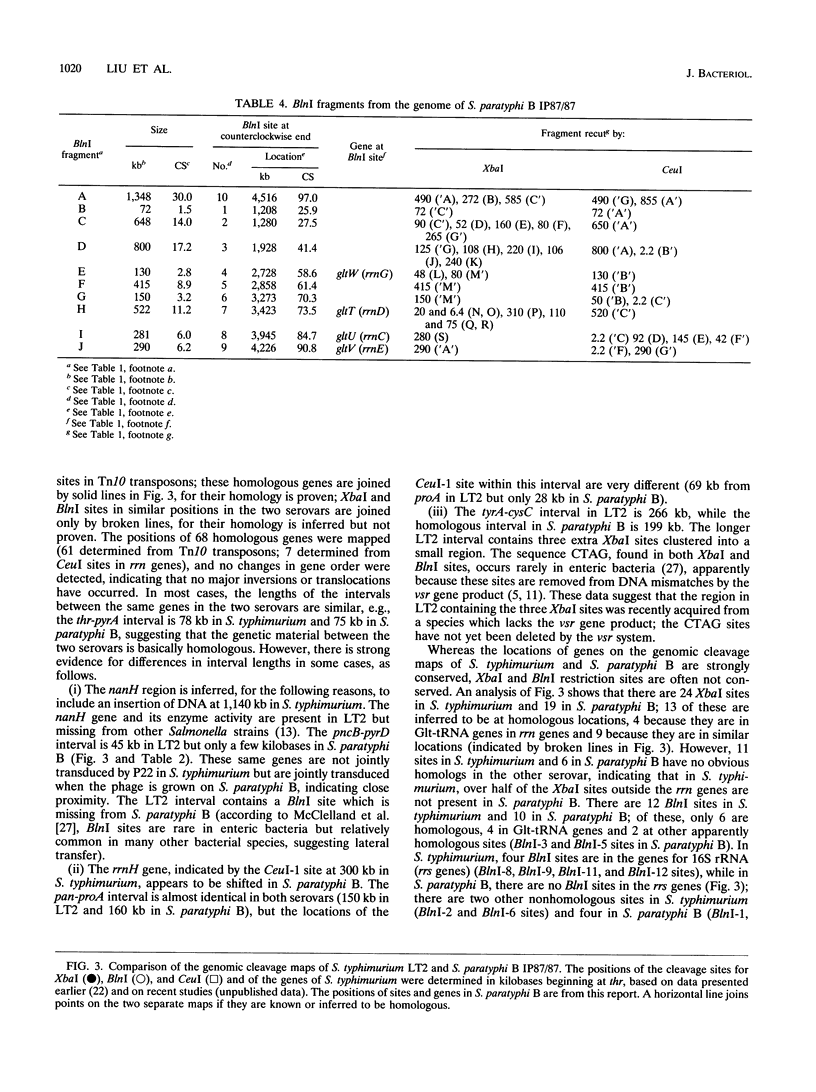
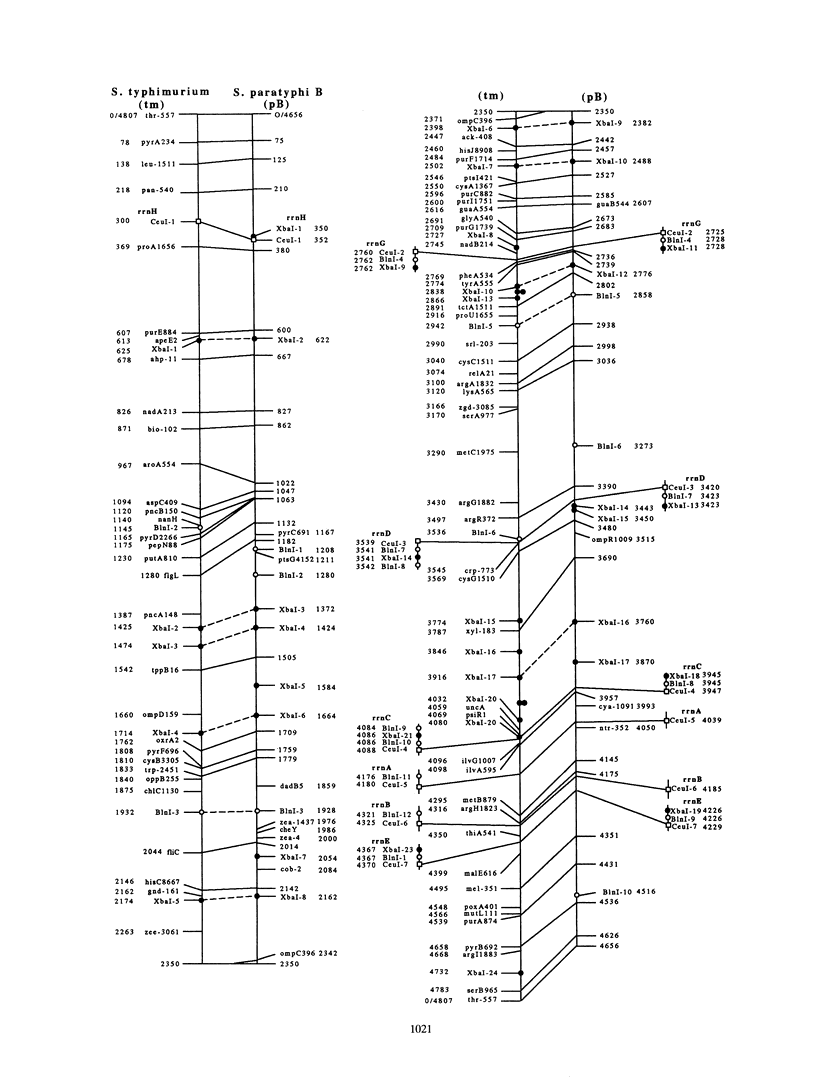
The genomic cleavage map of Salmonella paratyphi B was determined through digestion with endonucleases and separation of the fragments by pulsed-field gel electrophoresis. The chromosome has 19 XbaI sites, 10 BlnI sites, and 7 CeuI sites. The fragments were arranged in order through excision of fragments from the gel, redigestion with a second enzyme, end labelling with 32P, and reelectrophoresis. Tn10 transposons inserted in 61 different genes of S. typhimurium LT2 were transduced by use of bacteriophage P22 into S. paratyphi B. The locations of Tn10 insertions on the chromosome of S. paratyphi B were determined by use of XbaI and BlnI sites in Tn10, revealing the positions of genes with Tn10 insertions in S. paratyphi B. All seven CeuI sites (in rrl genes for 23S rRNA) and most of the XbaI and BlnI sites in rrn genes for Glt-tRNA are conserved, but only about half of the XbaI and BlnI sites outside rrn genes are conserved. Gene order is identical in the 68 genes that we could compare between S. paratyphi B and S. typhimurium LT2, and the lengths of intervals between the genes are often the same, but there are several instances of differences in interval lengths, indicating that insertions or deletions of DNA have occurred during the evolutionary divergence of these bacteria.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R. M., Kearney G. M., Nicholson P., Blair A. L., Porter R. C., Crichton P. B. Types of Salmonella paratyphi B and their phylogenetic significance. J Med Microbiol. 1988 Aug;26(4):285–293. doi: 10.1099/00222615-26-4-285. [DOI] [PubMed] [Google Scholar]
- Beltran P., Plock S. A., Smith N. H., Whittam T. S., Old D. C., Selander R. K. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J Gen Microbiol. 1991 Mar;137(3):601–606. doi: 10.1099/00221287-137-3-601. [DOI] [PubMed] [Google Scholar]
- Bhagwat A. S., McClelland M. DNA mismatch correction by Very Short Patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 1992 Apr 11;20(7):1663–1668. doi: 10.1093/nar/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Buvinger W. E., Lampel K. A., Bojanowski R. J., Riley M. Location and analysis of nucleotide sequences at one end of a putative lac transposon in the Escherichia coli chromosome. J Bacteriol. 1984 Aug;159(2):618–623. doi: 10.1128/jb.159.2.618-623.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casse F., Pascal M. C., Chippaux M. Comparison between the chromosomal maps of Escherichia coli and Salmonella typhimurium. Length of the inverted segment in the trp region. Mol Gen Genet. 1973 Aug 17;124(3):253–257. doi: 10.1007/BF00293096. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Ewing W. H., Falkow S. Molecular relationships among the Salmonelleae. J Bacteriol. 1973 Jul;115(1):307–315. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A., Turmel M., Lemieux C. A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr Genet. 1991 Jan;19(1):43–47. doi: 10.1007/BF00362086. [DOI] [PubMed] [Google Scholar]
- Hennecke F., Kolmar H., Bründl K., Fritz H. J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991 Oct 24;353(6346):776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- Hoyer L. L., Hamilton A. C., Steenbergen S. M., Vimr E. R. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol. 1992 Apr;6(7):873–884. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- KAUFFMANN F. Zur Differential diagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. Z Hyg Infektionskr. 1955;141(6):546–550. [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel K. A., Riley M. Discontinuity of homology of Escherichia coli and Salmonella typhimurium DNA in the lac region. Mol Gen Genet. 1982;186(1):82–86. doi: 10.1007/BF00422916. [DOI] [PubMed] [Google Scholar]
- Le Minor L. Typing of Salmonella species. Eur J Clin Microbiol Infect Dis. 1988 Apr;7(2):214–218. doi: 10.1007/BF01963091. [DOI] [PubMed] [Google Scholar]
- Le Minor L., Véron M., Popoff M. Proposition pour une nomenclature des Salmonella. Ann Microbiol (Paris) 1982 Sep-Oct;133(2):245–254. [PubMed] [Google Scholar]
- Lehner A. F., Harvey S., Hill C. W. Mapping and spacer identification of rRNA operons of Salmonella typhimurium. J Bacteriol. 1984 Nov;160(2):682–686. doi: 10.1128/jb.160.2.682-686.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol Microbiol. 1993 Nov;10(3):655–664. doi: 10.1111/j.1365-2958.1993.tb00937.x. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993 Jul;175(13):4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Sanderson K. E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992 Mar;174(5):1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K., Selander R. K. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992 Nov;174(21):6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff M. Y., Bockemühl J., McWhorter-Murlin A. Supplement 1991 (no. 35) to the Kauffmann-White scheme. Res Microbiol. 1992 Oct;143(8):807–811. doi: 10.1016/0923-2508(92)90109-2. [DOI] [PubMed] [Google Scholar]
- Popoff M. Y., Miras I., Coynault C., Lasselin C., Pardon P. Molecular relationships between virulence plasmids of Salmonella serotypes typhimurium and dublin and large plasmids of other Salmonella serotypes. Ann Microbiol (Paris) 1984 May-Jun;135A(3):389–398. doi: 10.1016/s0769-2609(84)80080-0. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Genetic relatedness in the family Enterobacteriaceae. Annu Rev Microbiol. 1976;30:327–349. doi: 10.1146/annurev.mi.30.100176.001551. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Hall C. A. F-prime factors of Salmonella typhimurium and an inversion between S. typhimurium and Escherichia coli. Genetics. 1970 Feb;64(2):215–228. doi: 10.1093/genetics/64.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H., Backhaus H. The origin of DNA in transducing particles in P22-mutants with increased transduction-frequencies (HT-mutants). Mol Gen Genet. 1973 Jan 24;120(2):181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Beltran P., Smith N. H., Barker R. M., Crichton P. B., Old D. C., Musser J. M., Whittam T. S. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect Immun. 1990 Jun;58(6):1891–1901. doi: 10.1128/iai.58.6.1891-1901.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992 Mar;174(5):1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]