Abstract
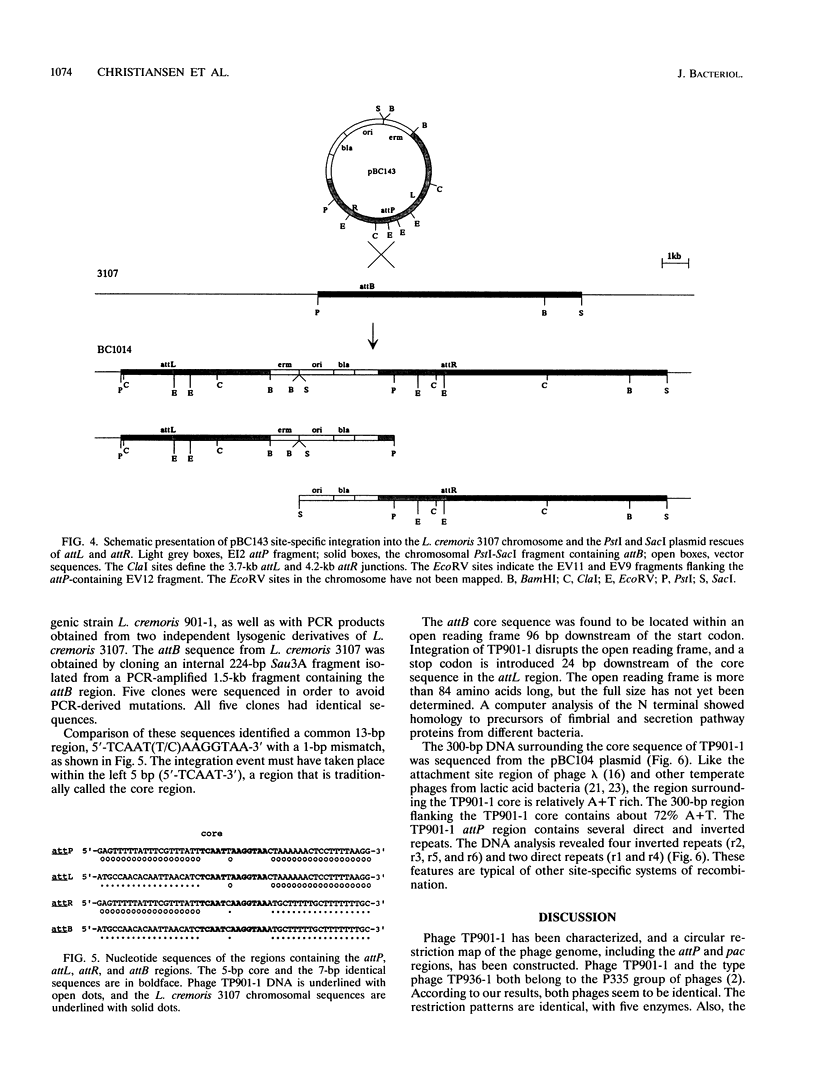
The temperate lactococcal phage TP901-1, induced by UV light from Lactococcus lactis subsp. cremoris 901-1, was characterized. The restriction map was found to be circular, and the packaging of TP901-1 DNA was concluded to occur by a headful mechanism. The pac region was localized on the 38.4-kb phage genome. TP901-1 belongs to the class of P335 phages (V. Braun, S. Hertwig, H. Neve, A. Geis, and M. Teuber, J. Gen. Microbiol. 135:2551-2560, 1989). Evidence is presented that the phages TP936-1 (V. Braun, S. Hertwig, H. Neve, A. Geis, and M. Teuber, J. Gen. Microbiol. 135:2551-2560, 1989) and C3-T1 (A. W. Jarvis, V. R. Parker, and M. B. Bianchin, Can. J. Microbiol. 38:398-404, 1992) are very closely related to or are identical to TP901-1. The lytically propagated TP901-1 phages were able to lysogenize both indicator strains Lactococcus cremoris 3107 and Wg2. Lysogenization resulted in site-specific integration of the phage genome into the bacterial chromosome. Only one chromosomal attB site was found in 20 independent lysogens. The attP region of TP901-1 and the attL and attR regions were cloned and sequenced. The results showed a core region of only 5 bp, in which the recombination occurs, followed after a 1-bp mismatch by a 7-bp identical region, TCAAT(T/C)AAGGTAA. This result was further verified by sequencing of the attB region obtained by PCR. An integration vector was constructed with the 6.5-kb EcoRI fragment from TP901-1 containing attP. This vector also functions in the plasmid-free strains, MG1363 and LM0230 with only one specific attB site, strongly indicating a more general use of the TP901-1-based integration vector in lactococci.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Campbell A. M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992 Dec;174(23):7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S., Huang W. M., Hayden M., Parr R. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987 Apr 5;194(3):411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- Coleman D., Knights J., Russell R., Shanley D., Birkbeck T. H., Dougan G., Charles I. Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage phi 13 occurs by site- and orientation-specific integration of the phi 13 genome. Mol Microbiol. 1991 Apr;5(4):933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x. [DOI] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977 Apr;130(1):257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J., Davies F. L. Prophage-Cured Derivatives of Streptococcus lactis and Streptococcus cremoris. Appl Environ Microbiol. 1980 Nov;40(5):964–966. doi: 10.1128/aem.40.5.964-966.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M. A., Scocca J. J. Location of the host attachment site for phage HPl within a cluster of Haemophilus influenzae tRNA genes. Nucleic Acids Res. 1990 Sep 11;18(17):5305–5305. doi: 10.1093/nar/18.17.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmidevi G., Davidson B. E., Hillier A. J. Circular Permutation of the Genome of a Temperate Bacteriophage from Streptococcus cremoris BK5. Appl Environ Microbiol. 1988 Apr;54(4):1039–1045. doi: 10.1128/aem.54.4.1039-1045.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmidevi G., Davidson B. E., Hillier A. J. Molecular characterization of promoters of the Lactococcus lactis subsp. cremoris temperate bacteriophage BK5-T and identification of a phage gene implicated in the regulation of promoter activity. Appl Environ Microbiol. 1990 Apr;56(4):934–942. doi: 10.1128/aem.56.4.934-942.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. J Bacteriol. 1988 May;170(5):2409–2411. doi: 10.1128/jb.170.5.2409-2411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Pascopella L., Jacobs W. R., Jr, Hatfull G. F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Lillehaug D., Birkeland N. K. Characterization of genetic elements required for site-specific integration of the temperate lactococcal bacteriophage phi LC3 and construction of integration-negative phi LC3 mutants. J Bacteriol. 1993 Mar;175(6):1745–1755. doi: 10.1128/jb.175.6.1745-1755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehaug D., Lindqvist B., Birkeland N. K. Characterization of phiLC3, a Lactococcus lactis subsp. cremoris temperature bacteriophage with cohesive single-stranded DNA ends. Appl Environ Microbiol. 1991 Nov;57(11):3206–3211. doi: 10.1128/aem.57.11.3206-3211.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya R. R., Fremaux C., De Antoni G. L., Klaenhammer T. R. Site-specific integration of the temperate bacteriophage phi adh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J Bacteriol. 1992 Sep;174(17):5584–5592. doi: 10.1128/jb.174.17.5584-5592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989 Mar 11;17(5):1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Goodman S. D., Scocca J. J. Nucleotide sequences and properties of the sites involved in lysogenic insertion of the bacteriophage HP1c1 genome into the Haemophilus influenzae chromosome. J Bacteriol. 1987 Jan;169(1):238–246. doi: 10.1128/jb.169.1.238-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]