Abstract
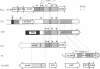
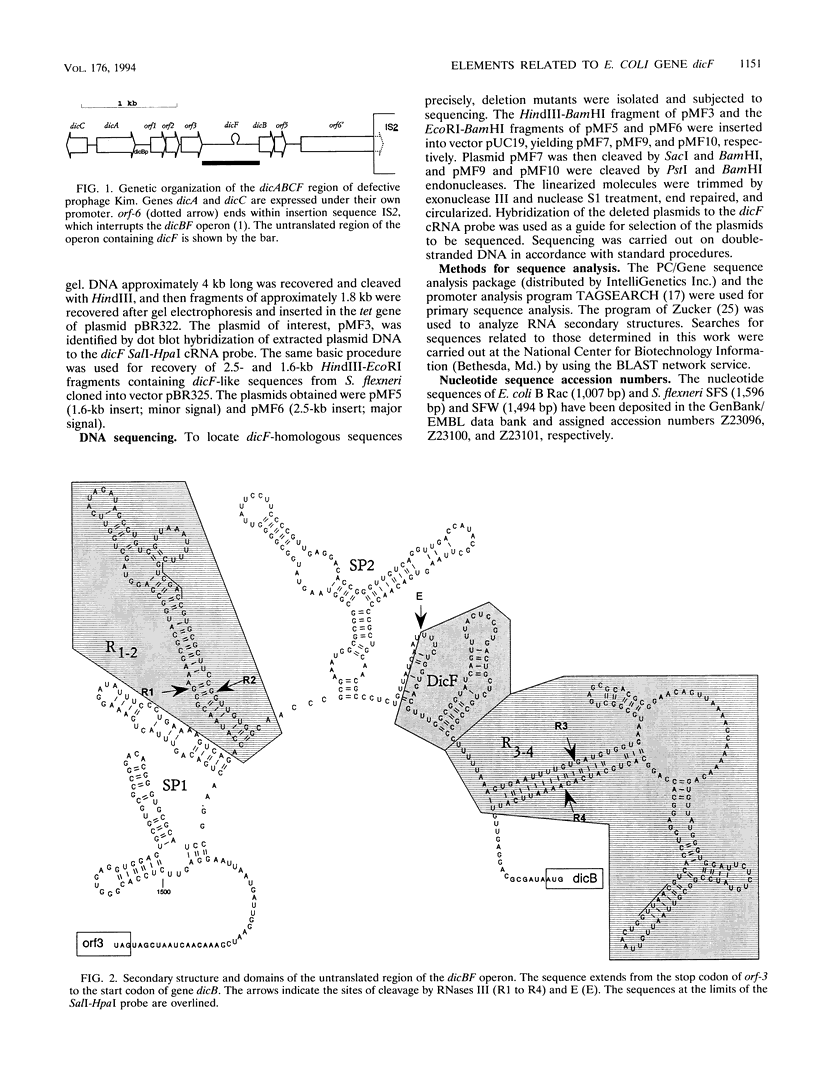
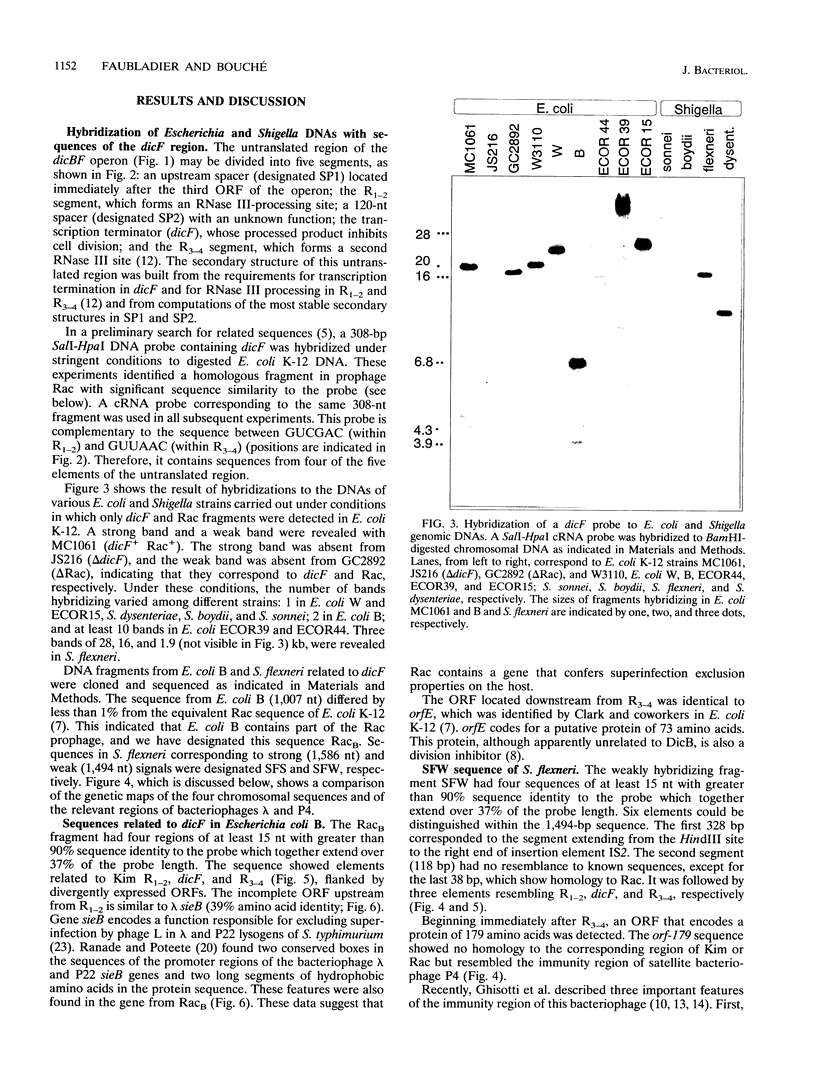
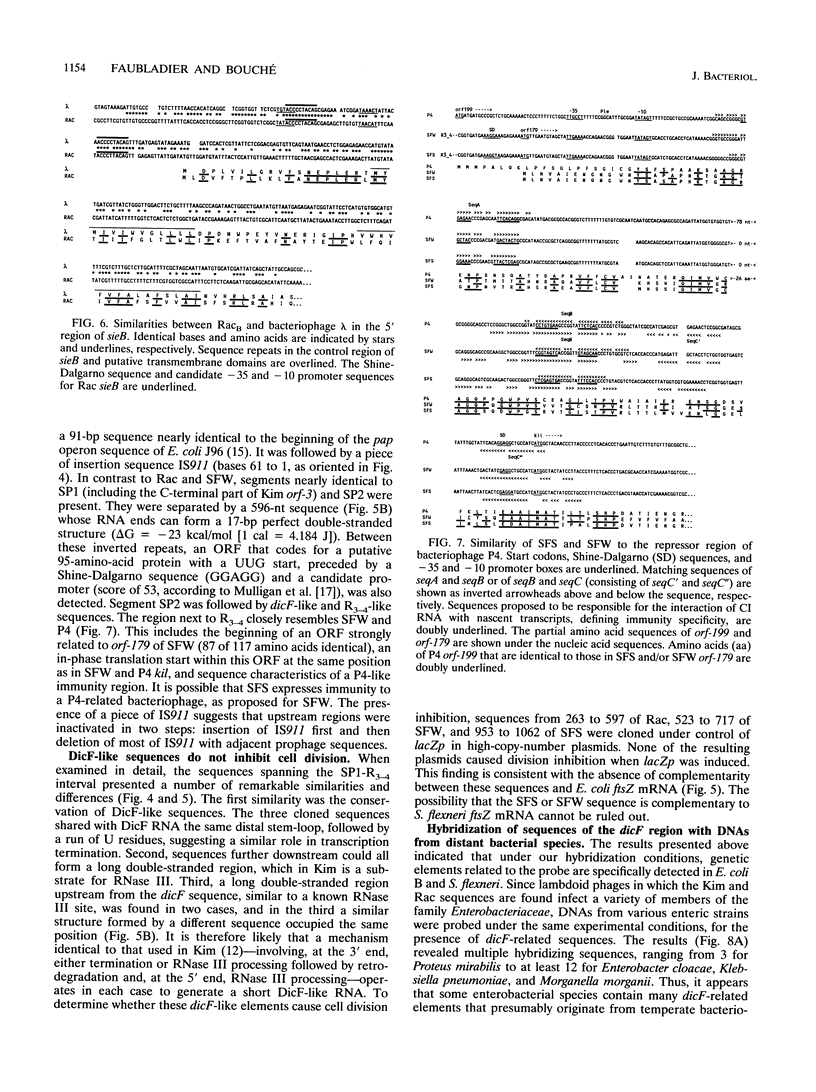
The genomes of various eubacteria were analyzed by Southern blot hybridization to detect sequences related to the segment of the defective lambdoid prophage Kim which encodes DicF RNA, an antisense inhibitor of cell division gene ftsZ in Escherichia coli K-12. Among the homologous sequences found, one fragment from E. coli B, similar to a piece of Rac prophage, and two fragments from Shigella flexneri were cloned and sequenced. dicF-like elements similar to transcriptional terminators were found in each sequence, but unlike dicF these had no effect on division in E. coli K-12. Like dicF, these sequences are flanked by secondary structures which form potential sites for RNase III recognition. Coding sequences located upstream from the dicF-like feature in E. coli B are related to gene sieB of bacteriophage lambda, while sequences downstream of the S. flexneri elements are similar to the immunity region of satellite bacteriophage P4. Under hybridization conditions in which only strong sequence homologies were detected in E. coli B and S. flexneri, the genomes of a large variety of microorganisms, including some gram-positive bacteria, hybridized to the dicF probe. Our results suggest that dicF and its flanking regions are markers of a widespread family of prophage-like elements of different origins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barcellini-Couget S., Galucci P., Bouché J. P. Escherichia coli dicB operon includes a truncated IS2 element. Nucleic Acids Res. 1988 Nov 11;16(21):10388–10388. doi: 10.1093/nar/16.21.10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché J. P., Gélugne J. P., Louarn J., Louarn J. M., Kaiser K. Relationships between the physical and genetic maps of a 470 x 10(3) base-pair region around the terminus of Escherichia coli K12 DNA replication. J Mol Biol. 1982 Jan 5;154(1):21–32. doi: 10.1016/0022-2836(82)90414-4. [DOI] [PubMed] [Google Scholar]
- Béjar S., Bouché F., Bouché J. P. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol Gen Genet. 1988 Apr;212(1):11–19. doi: 10.1007/BF00322439. [DOI] [PubMed] [Google Scholar]
- Béjar S., Bouché J. P. A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol Gen Genet. 1985;201(2):146–150. doi: 10.1007/BF00425651. [DOI] [PubMed] [Google Scholar]
- Dehó G., Zangrossi S., Sabbattini P., Sironi G., Ghisotti D. Bacteriophage P4 immunity controlled by small RNAs via transcription termination. Mol Microbiol. 1992 Nov;6(22):3415–3425. doi: 10.1111/j.1365-2958.1992.tb02209.x. [DOI] [PubMed] [Google Scholar]
- Espion D., Kaiser K., Dambly-Chaudiere C. A third defective lambdoid prophage of Escherichia coli K12 defined by the lambda derivative, lambdaqin111. J Mol Biol. 1983 Nov 5;170(3):611–633. doi: 10.1016/s0022-2836(83)80124-7. [DOI] [PubMed] [Google Scholar]
- Faubladier M., Cam K., Bouché J. P. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J Mol Biol. 1990 Apr 5;212(3):461–471. doi: 10.1016/0022-2836(90)90325-G. [DOI] [PubMed] [Google Scholar]
- Ghisotti D., Chiaramonte R., Forti F., Zangrossi S., Sironi G., Dehò G. Genetic analysis of the immunity region of phage-plasmid P4. Mol Microbiol. 1992 Nov;6(22):3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Dehò G., Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol Rev. 1993 Sep;57(3):683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund B. I., Tennent J. M., Garcia E., Hamers A., Båga M., Lindberg F., Gaastra W., Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992 Aug;6(16):2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls R. D., Hill A. V., Clegg J. B., Higgs D. R. Direct cloning of specific genomic DNA sequences in plasmid libraries following fragment enrichment. Nucleic Acids Res. 1985 Nov 11;13(21):7569–7578. doi: 10.1093/nar/13.21.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Ranade K., Poteete A. R. Superinfection exclusion (sieB) genes of bacteriophages P22 and lambda. J Bacteriol. 1993 Aug;175(15):4712–4718. doi: 10.1128/jb.175.15.4712-4718.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. A., Dykhuizen D. E., DuBose R. F., Green L., Mutangadura-Mhlanga T., Wolczyk D. F., Hartl D. L. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987 Jan;115(1):51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susskind M. M., Botstein D. Superinfection exclusion by lambda prophage in lysogens of Salmonella typhimurium. Virology. 1980 Jan 15;100(1):212–216. doi: 10.1016/0042-6822(80)90571-1. [DOI] [PubMed] [Google Scholar]
- Tétart F., Bouché J. P. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol. 1992 Mar;6(5):615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]