Abstract
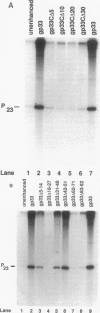
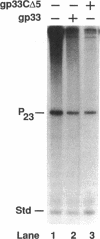
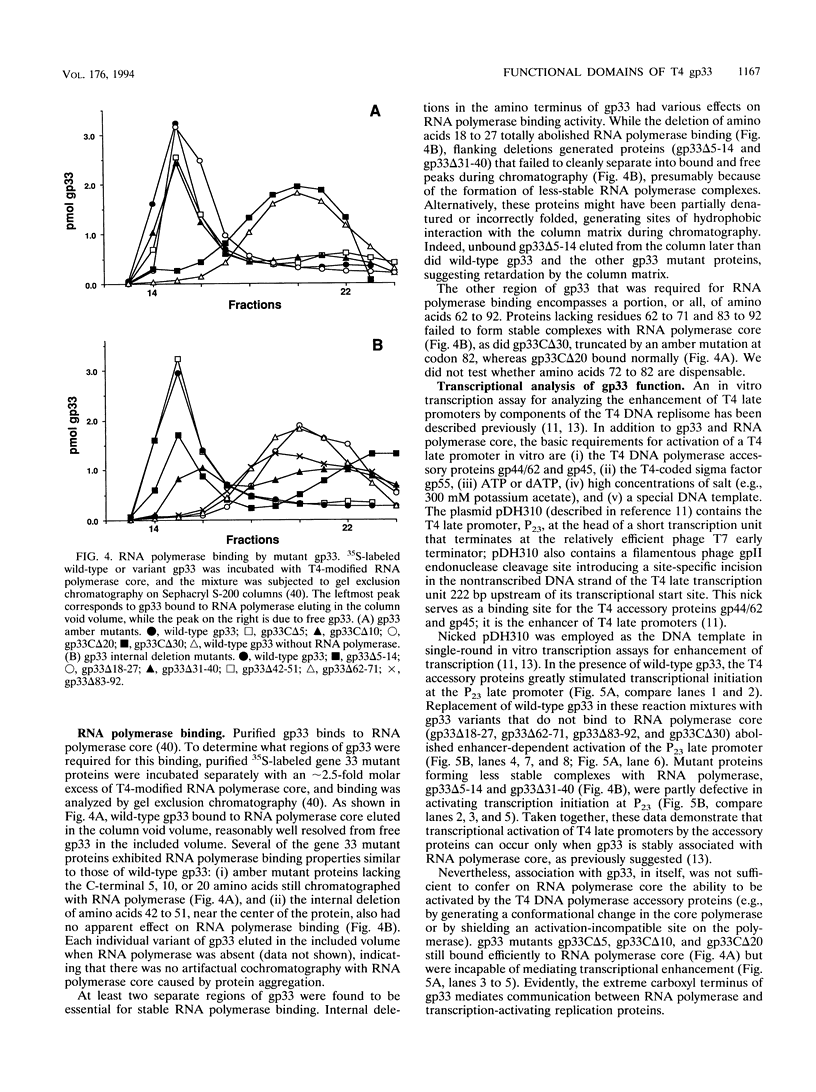
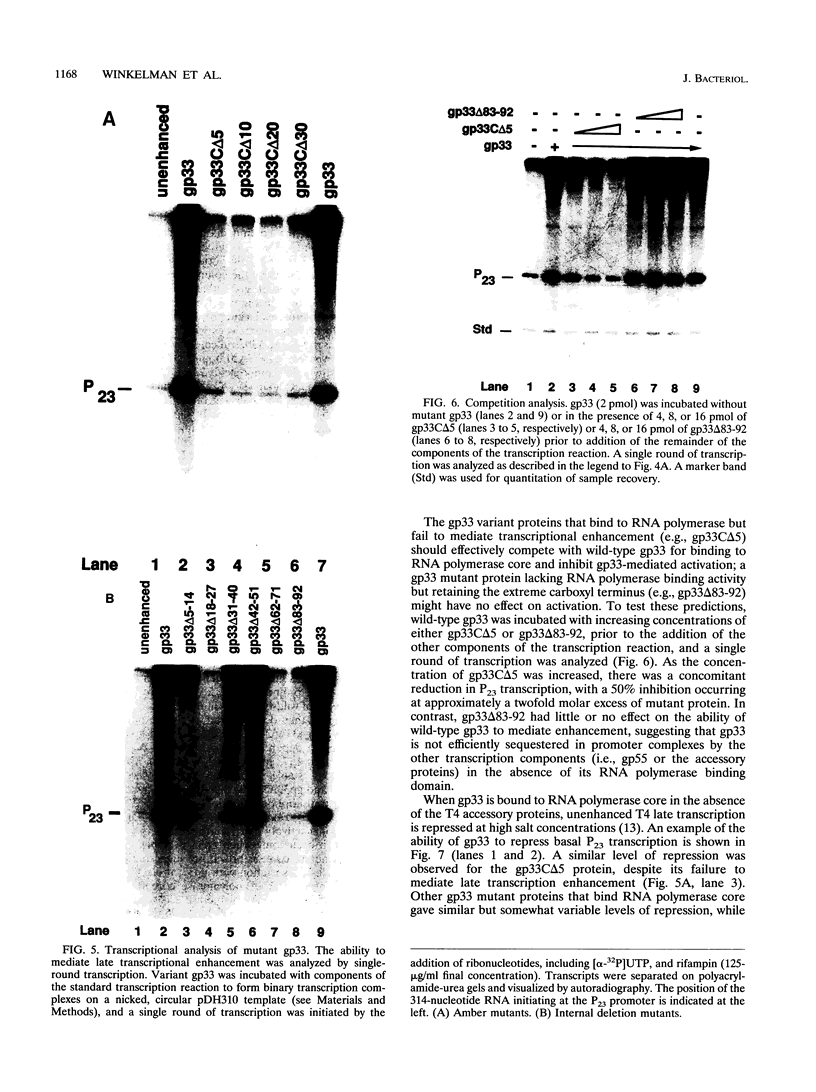
The bacteriophage T4 gene 33 encodes a small, acidic RNA polymerase-binding protein that mediates enhancement of transcriptional initiation at T4 late promoters by the T4 DNA replication accessory proteins. A set of nested deletions in the gene 33 open reading frame was constructed by oligonucleotide site-directed mutagenesis. The resulting variant gene 33 proteins were radiolabeled during overexpression employing a T7 RNA polymerase-based system and substantially purified. Each variant was analyzed for three properties of gp33: RNA polymerase binding activity, ability to mediate enhancer-dependent transcriptional activation, and repression of unenhanced transcription. Two separate regions of gp33 were required to form stable complexes with RNA polymerase, whereas the extreme carboxyl terminus of gp33 was essential for mediating late gene activation. Variant gene 33 proteins lacking the carboxyl terminus nevertheless repressed nonenhanced transcription, demonstrating that the functional domains required for transcriptional activation and repression of unenhanced transcription are separable. The possible roles of gp33 in mediating late gene expression are discussed in the light of the identification of these functional domains.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartholomew B., Kassavetis G. A., Braun B. R., Geiduschek E. P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990 Jul;9(7):2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F. Host mutant (tabD)-induced inhibition of bacteriophage T4 late transcription. II. Genetic characterization of mutants. J Mol Biol. 1975 Aug 25;96(4):601–624. doi: 10.1016/0022-2836(75)90141-2. [DOI] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F., Takahashi H. Host mutant (tabD)-induced inhibition of bacteriophage T4 late transcription. I. Isolation and phenotypic characterization of the mutants. J Mol Biol. 1975 Aug 25;96(4):579–600. doi: 10.1016/0022-2836(75)90140-0. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Ebright R. H. Transcription activation at Class I CAP-dependent promoters. Mol Microbiol. 1993 May;8(5):797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J., 3rd, Sayre M. H., Tschochner H., Kornberg R. D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991 Apr 4;350(6317):436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P. Regulation of expression of the late genes of bacteriophage T4. Annu Rev Genet. 1991;25:437–460. doi: 10.1146/annurev.ge.25.120191.002253. [DOI] [PubMed] [Google Scholar]
- Hahn S., Kruse U., Rüger W. The region of phage T4 genes 34, 33 and 59: primary structures and organization on the genome. Nucleic Acids Res. 1986 Dec 9;14(23):9311–9327. doi: 10.1093/nar/14.23.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward R. S., Igarashi K., Ishihama A. Functional specialization within the alpha-subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991 Sep 5;221(1):23–29. doi: 10.1016/0022-2836(91)80197-3. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Williams K. P., Kassavetis G. A., Geiduschek E. P. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990 May 4;248(4955):573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- Heyduk T., Lee J. C., Ebright Y. W., Blatter E. E., Zhou Y., Ebright R. H. CAP interacts with RNA polymerase in solution in the absence of promoter DNA. Nature. 1993 Aug 5;364(6437):548–549. doi: 10.1038/364548a0. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R. Polypeptide bound to the host RNA polymerase is specified by T4 control gene 33. Nat New Biol. 1973 Aug 1;244(135):137–140. doi: 10.1038/newbio244137a0. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Ji T. H. Bifunctional reagents. Methods Enzymol. 1983;91:580–609. doi: 10.1016/s0076-6879(83)91053-4. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990 Jun 29;61(7):1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Mencía M., Salas M., Rojo F. Residues of the Bacillus subtilis phage phi 29 transcriptional activator required both to interact with RNA polymerase and to activate transcription. J Mol Biol. 1993 Oct 20;233(4):695–704. doi: 10.1006/jmbi.1993.1546. [DOI] [PubMed] [Google Scholar]
- Orsini G., Brody E. N. Phage T4 DNA codes for two distinct 10-kDa proteins which strongly bind to RNA polymerase. Virology. 1988 Feb;162(2):397–405. doi: 10.1016/0042-6822(88)90480-1. [DOI] [PubMed] [Google Scholar]
- Park C. S., Wu F. Y., Wu C. W. Molecular mechanism of promoter selection in gene transcription. II. Kinetic evidence for promoter search by a one-dimensional diffusion of RNA polymerase molecule along the DNA template. J Biol Chem. 1982 Jun 25;257(12):6950–6956. [PubMed] [Google Scholar]
- Piperno J. R., Kallen R. G., Alberta B. M. Analysis of a T4 DNA replication protein complex. Studies of the DNA recognition site for T4 gene 44/62 and 45 protein-catalyzed ATP hydrolysis. J Biol Chem. 1978 Jul 25;253(14):5180–5185. [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Ricchetti M., Metzger W., Heumann H. One-dimensional diffusion of Escherichia coli DNA-dependent RNA polymerase: a mechanism to facilitate promoter location. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4610–4614. doi: 10.1073/pnas.85.13.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Williams K. P., Müller R., Rüger W., Geiduschek E. P. Overproduced bacteriophage T4 gene 33 protein binds RNA polymerase. J Bacteriol. 1989 Jun;171(6):3579–3582. doi: 10.1128/jb.171.6.3579-3582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Busby S., Ebright R. H. Identification of the functional subunit of a dimeric transcription activator protein by use of oriented heterodimers. Cell. 1993 Apr 23;73(2):375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]