Abstract
Translocation of ricin A chain to the cytosol has been proposed to take place from the endoplasmic reticulum (ER), but attempts to visualize ricin in this organelle have failed. Here we modified ricin A chain to contain a tyrosine sulfation site alone or in combination with N-glycosylation sites. When reconstituted with ricin B chain and incubated with cells in the presence of Na235SO4, the modified A chains were labeled. The labeling was prevented by brefeldin A and ilimaquinone, and it appears to take place in the Golgi apparatus. This method allows selective labeling of ricin molecules that have already been transported retrograde to this organelle. A chain containing C-terminal N-glycosylation sites became core glycosylated, indicating retrograde transport to the ER. In part of the toxin molecules, the A chain was released from the B chain and translocated to the cytosol. The finding that glycosylated A chain was present in the cytosol indicates that translocation takes place after transport of the toxin to the ER.
Keywords: ricin, toxin, translocation, retrograde transport, sulfation
Ricin is a plant toxin that is extensively used in the construction of immunotoxins for targeted therapy of cancer and other diseases (1–3). The toxin consists of an enzymatically active A chain and a B chain with lectin properties (4). The A chain enters the cytosol and inactivates ribosomes by depurination of a single adenosine residue in 28S ribosomal RNA (5, 6). The B chain binds to galactose-containing surface receptors (7). The surface-bound toxin is slowly endocytosed (8). Part of the internalized ricin is transported back to the cell surface, part is delivered to lysosomes and degraded, and part is transported to the trans-Golgi network (9). Translocation of ricin A chain to the cytosol has been proposed to take place from the endoplasmic reticulum (ER; refs. 10–12). In fact, Shigella toxin, which has the same intracellular target as ricin (13, 14), has been visualized in the ER (15), and the related cholera and Escherichia coli heat labile toxins have C-terminal (K/H)DEL sequences that could act as ER retrieval signals (16). Pseudomonas aeruginosa exotoxin A has a C terminus that, after removal of the C-terminal lysine residue, ends in RDEL (16). Ricin does not contain such a sequence. However, addition of a KDEL sequence to the C terminus of ricin A chain was found to increase the toxic effect of whole ricin and, in particular, that of the free A chain (10, 12, 18).
To test if ricin is transported retrograde from the trans-Golgi network to the ER, we decided to use transfer of N-linked oligosaccharides onto free glycosylation sites in ricin A chain as a test for the presence of toxin in the ER. It has been found that only ≈5% of endocytosed ricin is located in the trans-Golgi network (19). It follows that if labeled toxin is added to the cells, there will be a serious background problem when one looks for toxin in the Golgi apparatus and in the ER. To reduce this background, we used tyrosine sulfation as a labeling procedure (20). Because this labeling takes place in the Golgi apparatus, only molecules that have already been transported retrograde to the Golgi complex will be labeled. Furthermore, because only the A chain carrying the sulfation site will be labeled, the B chain, which migrates at almost the same rate as the modified A chain, will not complicate the interpretation of the data. We here present evidence that sulfated ricin A chain is transported retrograde to the ER and then translocated to the cytosol.
MATERIALS AND METHODS
Materials and Buffers.
Na235SO4 was from Amersham. Rat monoclonal (9G10) anti-glucose regulated protein 94 (GRP94) was from StressGen Biotechnologies (Victoria, Canada). Affinity-purified rabbit anti-BIP, was obtained from Linda Hendershot (St. Jude’s Children Hospital, Memphis, TN). This antibody was found to cross-react with cytosolic heat shock protein 70 (HSP70). Mouse monoclonal (1D3) anti-protein disulfate isomerase (PDI) was obtained from Steven Fuller (European Molecular Biology Laboratory, Heidelberg, Germany). Anti-p58 was obtained from Jaakko Saraste (University of Bergen, Bergen, Norway). Anti-rab5 was obtained from Harald Stenmark (Institute for Cancer Research, Oslo). Hepes medium contained bicarbonate-free Eagle’s minimum essential medium buffered with 20 mM Hepes (N-2-hydroxethylpiperazine-N′-2-ethanesulfonic acid) to pH 7.4. PBS contained 140 mM NaCl and 10 mM Na2HPO4 (pH 7.2). Lysis buffer consisted of PBS (pH 7.2) containing 1 mM EDTA, 1% Triton X-100, 200 units/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Rabbit anti-ricin was obtained by standard immunization. Endoglycosidase H (endo H) and endoglycosidase F (PNGase F) were from New England Biolabs.
Cell Culture.
Cells were maintained and propagated under standard conditions (5% CO2 in Eagle’s minimal essential medium containing 5% fetal calf serum). Two days before the experiment, the cells were seeded into 12- or 24-well Costar plates at a density of 105 and 5 × 104 cells per well, respectively.
Plasmids.
To form ricin A-sulf-1, a C-terminal extension was introduced by PCR using wild-type ricin A chain cDNA (21) as template and 5′-GGATCCTATCAAGATGGGTATTCGTAGTCTTCTGCGCTGCTTGGTGGAGGTGCGCATCTATACACC-3′ as the reverse primer. To form ricin A-sulf-2, ricin A-sulf-1 was used as template, and 5′-CGTCGGATCCCGGGCTATCACTGGGATGTGTTATTTTTGGTGCCGTTAGATGGGTATTCGTAGTCTTCTGCGC-3′ was used as the reverse primer. In both cases 5′-GCAATCACTCATCTTTTCACTGATGTTC-3′ was used as the forward primer. The product was cut with BglII and BamHI and cloned into pRA (21) that had been cut with the same enzymes and dephosphorylated. From the plasmids obtained, cDNA for the modified A chains were cut out with NcoI and EcoRI and cloned into pMAL-cN which had been cut partially with NcoI and to completion with EcoRI. pMAL-cN was obtained by cutting pMAL-c (New England Biolabs) with StuI and ligating in an NcoI linker: 5′-GCCATGGC-3′.
Measurement of Cytotoxicity of Endocytosed Toxin.
Vero cells were incubated for 4 h in Hepes medium without leucine with increasing amounts of toxin. The cells were then transferred to Hepes medium containing 1 μCi/ml [3H]leucine (1 Ci = 37 GBq) and no unlabeled leucine and were incubated for 20 min at 37°C. The cells were then extracted with 5% trichloroacetic acid for 10 min, followed by a brief wash in 5% trichloroacetic acid, and subsequently dissolved in 0.1 M KOH, and the cell-associated radioactivity was measured.
Glycosidase Treatment.
Vero cells were incubated with ricin containing A-sulf-2, lysed, and immunoprecipitated with immobilized anti-ricin. The beads were heated at 100°C for 10 min in denaturing buffer (0.5% SDS/1% mercaptoethanol). Digestion was carried out with Endo H or PNGase F for 2 h at 37°C. The reaction was terminated by the addition of concentrated SDS sample buffer and boiling for 5 min.
Purification of Recombinant Proteins and Reconstitution with Ricin B Chain.
After induction with isopropyl β-d-thiogalactoside, bacteria were harvested, washed with PBS, resuspended in column buffer (20 mM Tris·Cl, pH 7.5/200 mM NaCl/0.1 mM PMSF) and sonicated. After centrifugation at 12,000 × g for 10 min, the clear supernatant was applied to a column with amylose resin. The fusion proteins were eluted with column buffer containing 10 mM maltose. Free ricin A chain was cleaved off with factor Xa, mixed with the ricin B chain and dialyzed extensively against PBS to remove reducing agents.
Permeabilization of Cells with Streptolysin O (SLO).
Vero cells were incubated with 35SO42− and reconstituted ricin for 4 h. Then 10 mM N-ethylmaleimide was added, and the cells were incubated further for 10 min and then washed twice with PBS containing 0.1 M lactose. The cells were then placed on ice, and 2 μg/ml SLO preactivated with 10 mM MesNA in Hepes medium (pH 7.5) was added. After 10 min, the cells were washed briefly to remove unbound toxin and incubated for 10 min at 37°C for permeabilization to occur. Subsequently, the cells were kept on ice for an additional 30 min to allow components of the cytosol to diffuse into the buffer. Finally, the cells were scraped from the dish, centrifuged, and lysed with 0.5% Triton X-100. The lysate and cytosol were exposed to immobilized anti-ricin antibodies, and the adsorbed material was subjected to nonreducing SDS/PAGE.
SDS/PAGE.
SDS/PAGE was carried out as described (22) in the absence or presence of 2-mercaptoethanol, as indicated. The gels were fixed in 4% acetic acid/27% methanol for 30 min and then, in the case of proteins labeled with Na235SO4, treated with 1 M sodium salicylate (pH 5.8) in 2% glycerol for 30 min. Dried gels were exposed to Kodak XAR-5 films in the absence of intensifying screens at −80°C for fluorography.
Immunoblots.
The samples were subjected to reducing SDS/PAGE in a 12% gel, and proteins were transferred to a poly(vinylidene difluoride) membrane (Immobilon-P, Millipore). After blocking with 5% milk in PBS for 1 h, the membranes were incubated for 1 h with rabbit anti-Rab 5 antisera (1:1000), rabbit anti-BIP antibody (1 μg/ml), rabbit anti-p58 antisera (1:500), rat monoclonal anti-GRP94 (1:1000), or mouse monoclonal anti-PDI antibody (1:10). Then the blots were incubated with a second antibody labeled with alkaline phosphatase (Promega), and proteins were visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium solution according to the manufacturer’s instruction.
RESULTS
Ricin A Chain, Carrying a C-Terminal Tyrosine Sulfation Site and Reconstituted with Ricin B Chain, Is Toxic to Cells.
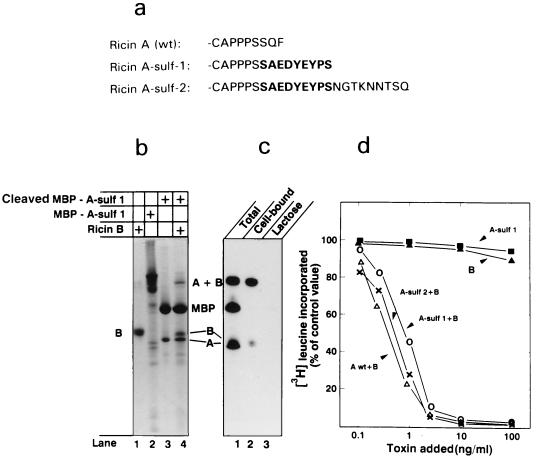
Ricin does not contain an endogenous sulfation site, and we therefore engineered one into the C terminus of ricin A chain. For this purpose, we used a nonapeptide that is functioning as a tyrosine sulfation site in rat cholecystokinin precursor (ref. 20; Fig. 1a). In one construct (ricin A-sulf-2), we also added three partly overlapping N-glycosylation sites. The constructs were made as fusion proteins with MBP (Fig. 1b, lane 2) to allow purification on a column of amylose resin. A factor Xa cleavage site was inserted at the junction between the two proteins, and after purification, the modified A chain was cleaved off with factor Xa (Fig. 1b, lane 3). When ricin B chain (Fig. 1b, lane 1) was added and the mixture was dialyzed to remove reducing agents and thereby allow formation of an interchain disulfide bond, holotoxin (A + B) was formed (Fig. 1b, lane 4). Only part of the modified A chain was able to form a disulfide-linked heterodimer with the B chain. MBP obtained upon cleavage of the fusion protein did not interfere with the association and was therefore in most cases not removed.
Figure 1.
Formation of modified ricin A chains and their reconstitution with the B chain to form holotoxin. (a) The C-terminal end of wild-type ricin A chain and A chain modified to contain sulfation and glycosylation sites. The C-terminal nonapeptide of rat cholecystokinin precursor (indicated with boldface type) was added to the C-terminal end of ricin A chain to form ricin A-sulf-1. To form ricin A-sulf-2, a nonapeptide containing three partially overlapping N-glycosylation sites was added to the C terminus of ricin A-sulf-1. The constructs were produced as fusion proteins with maltose-binding protein (MBP) and purified on an amylose column. (b) Lanes: 1, ricin B chain; 2, uncleaved fusion protein containing ricin A-sulf-1; 3, the same as lane 2, but after cleavage with factor Xa; 4, reconstituted ricin obtained by dialyzing together material as in lanes 1 and 3. (c) Material as in b, lane 4, labeled with 125I (lane 1) was added to Vero cells at 4°C for 30 min in the absence (lane 2) and presence (lane 3) of 10 mM lactose. The cells were then washed and analyzed by SDS/PAGE under nonreducing conditions. (d) Increasing amounts of wild-type and reconstituted ricin or the free A and B chains were added to Vero cells and incubated for 4 h. Then the ability of the cells to incorporate [3H]leucine was measured.
When the reconstituted, but not purified, toxin was labeled with 125I and analyzed by SDS/PAGE, bands corresponding to the reconstituted toxin (A + B) as well as MBP and the free A and B chains were obtained (Fig. 1c, lane 1). When added to Vero cells at 4°C, only reconstituted ricin and free B chain, but not free A chain or MBP, were bound to the cells (Fig. 1c, lane 2). No binding was obtained in the presence of lactose (Fig. 1c, lane 3), which is a competitive inhibitor of the binding of ricin to cell surface binding sites (7).
Although the free A and B chains were not able to inhibit protein synthesis measured as incorporation of [3H]leucine in Vero cells, the reconstituted holotoxins were in both cases essentially as toxic as wild-type ricin (Fig. 1d). The data therefore demonstrate that ricin A chain carrying C-terminal sulfation and glycosylation sites reconstitutes with ricin B chain to form fully active toxin.
Ricin Containing a Tyrosine Sulfation Site Is Sulfated upon Incubation with Cells.
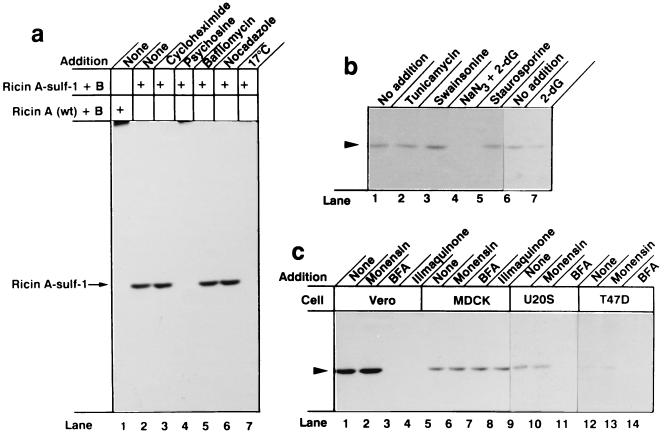
When cells were incubated with wild-type ricin in the presence of Na235SO4 and then lysed, and ricin was recovered with immobilized anti-ricin and analyzed by SDS/PAGE, there was, as expected, no labeling of the toxin (Fig. 2a, lane 1). However, when ricin containing A chain-sulf-1 was used, a distinct band migrating corresponding to the A chain was obtained (Fig. 2a, lane 2). Whereas cycloheximide, bafilomycin A1, and nocodazole, which inhibit cellular protein synthesis, vesicle acidification, and microtubule polymerization, respectively (23), did not inhibit the labeling (Fig. 2a, lanes 3, 5, and 6), psychosine (lane 4), which inhibits tyrosylprotein sulfotransferase (24), and incubation at 17°C (lane 7), which blocks retrograde transport of ricin to the Golgi complex (25), prevented the labeling. Psychosine is also known as an inhibitor of protein kinase C, but staurosporine, a strong inhibitor of protein kinase C (26) had no effect on the sulfation (Fig. 2b, lane 5).
Figure 2.
Sulfation of ricin A-sulf-1 upon incubation of reconstituted toxin with different cells. (a) Near confluent Vero cells growing in 5-cm2 dishes were washed twice with DMEM without sulfate and incubated with the same medium containing 100 μCi/ml Na235SO4 for 3 h. The indicated compounds were added, and after 30 min, 200 ng/ml reconstituted ricin containing ricin A-wt (lane 1) or ricin A-sulf-1 (lanes 2–7) was added and the incubation was continued for 4 h. Then the cells were washed with PBS and lysed, the nuclei were removed by centrifugation, and the clear supernatant was submitted to immunoprecipitation with rabbit anti-ricin antibodies immobilized on CNBr-Sepharose 4B. The adsorbed material was analyzed by SDS/PAGE under reducing conditions. The additions were as follows: lane 3, 20 μg/ml cycloheximide; lane 4, 20 μM psychosine; lane 5, 0.1 μM bafilomycin A1; lane 6, 30 μM nocodazole; lane 7, the incubation temperature was 17°C. (b) Conditions were as in a, lane 2, but the additions were as follows: lane 2, 1 μM tunicamycin; lane 3, 1 μg/ml swainsonine; lane 4, 10 mM NaN3 and 50 mM 2-deoxyglucose; lane 5, 0.1 μM staurosporil; lane 7, 50 mM 2-deoxyglucose. (c) Cells as indicated were treated as in b with the following additions: lanes 2, 6, 10, and 13, 10 μM monensin; lanes 3, 7, 11, and 14, 2 μg/ml brefeldin A; lanes 4 and 8, 30 μM ilimaquinone.
Tunicamycin and swainsonine, which interfere with protein glycosylation (23), did not inhibit the labeling (Fig. 2b, lanes 2 and 3), whereas depletion of ATP by incubation with 2-deoxyglucose and NaN3 (27) precluded labeling (lane 4). 2-Deoxyglucose alone, which does not strongly reduce the ATP level, had little inhibitory effect (Fig. 2b, lane 7).
Labeling occurred in all cell lines tested (Fig. 2c). In none of the cells did monensin inhibit the labeling (Fig. 2c, lanes 2, 6, 10, and 13), whereas brefeldin A (lanes 3, 7, 11, and 14) and ilimaquinone (lanes 4 and 8), which disrupt the Golgi apparatus by different mechanisms (28, 29), prevented labeling in those cells tested, except MDCK cells. In MDCK cells, the Golgi apparatus is not disrupted by brefeldin A (30). Brefeldin A and ilimaquinone protect those cells against ricin intoxication whose Golgi apparatus is disrupted by the drugs (31). MDCK cells are fully sensitive to ricin in the presence of brefeldin A (30). The data indicate that ricin containing a sulfation site is efficiently labeled with 35SO42− under conditions where retrograde transport to the Golgi apparatus occurs.
Cell-Mediated Glycosylation of Ricin A Chain with C-Terminal Glycosylation Sites.
Natural ricin A chain contains two N-linked oligosaccharide chains (32). Recombinant A chain produced in bacteria lacks these oligosaccharides, and we considered the possibility that if the recombinant toxin is transported retrograde to the ER, it may become glycosylated. However, we found no evidence for glycosylation of the sulfate-labeled A chain. This could mean that the relevant asparagine residues are not exposed on the folded A chain, and we therefore engineered three partly overlapping N-glycosylation sites onto the C terminus of ricin A-sulf-1 to yield ricin A-sulf-2 (Fig. 1a) in the hope that at least one of the sites would be sufficiently exposed for glycosylation to occur.
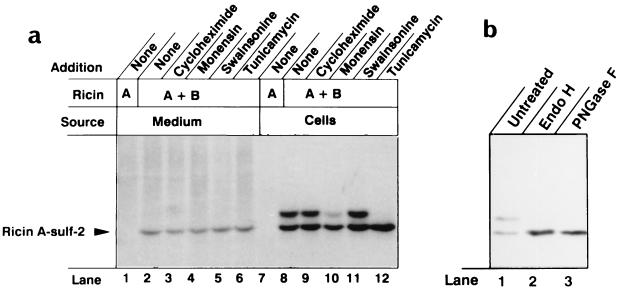
When the modified A chain was reconstituted with B chain and given to cells, two labeled bands appeared (Fig. 3a, lane 8). In the presence of tunicamycin, which inhibits core glycosylation of proteins in the ER (23), only the lighter band, migrating like unmodified ricin A-sulf-2 was observed (lane 12), indicating that the heavier band represents glycosylated molecules. At a comparatively high concentration (10 μM), monensin partly inhibited the formation of the heavier band (lane 10) in accordance with its ability to inhibit transport within the Golgi apparatus (33). We have shown that this concentration of monensin protects partially against ricin intoxication (27). Swainsonine, which inhibits processing of the attached oligosaccharide (23), did not inhibit the appearance of the heavier band (Fig. 3a, lane 11). Also nocodazole, which induces fragmentation of the Golgi apparatus into a large number of mini-Golgi apparatuses located adjacent to ER exit sites and in functional connection with the ER (34), did not inhibit the glycosylation (data not shown). After treatment with endo H, which removes from glycoproteins N-linked core oligosaccharide chains but not mature chains (35), and PNGase F, which removes both core and mature oligosaccharides, only the rapidly migrating form was observed (Fig. 3b, lanes 2 and 3).
Figure 3.
Sulfation and glycosylation of ricin A-sulf-2. (a) Vero cells were incubated with 35SO42− and ricin A-sulf-2 alone (lanes 1 and 7) or ricin A-sulf-2 reconstituted with ricin B chain (lanes 2–6 and 8–12). The immunoprecipitated material in the medium (lanes 1–6) and in the dissolved cells (lanes 7–12) was analyzed by SDS/PAGE under reducing conditions. The additions were as follows: lanes 3 and 9, 20 μg/ml cycloheximide; lanes 4 and 10, 10 μM monensin; lanes 5 and 11, 1 μg/ml swainsonine; lanes 6 and 12, 1 μM tunicamycin. (b) Vero cells incubated with 35SO42− and reconstituted ricin containing A-sulf-2 were lysed and immunoprecipitated with immobilized anti-ricin. The samples were either kept untreated or treated with Endo H and PNGase F, as indicated. Then sample buffer was added, and the samples were boiled for 5 min and submitted to SDS/PAGE under reducing conditions.
Part of the 35S-labeled A chain was found in the medium (Fig. 3a, lanes 2–6), indicating that it was exocytosed after being labeled in the Golgi apparatus. Only unglycosylated A chain was found in the medium, indicating that the sorting occurred at the level of the Golgi and that toxin that had once arrived in the ER was not transported out again. Also washing the cells with buffer containing lactose and mannose did not release toxin containing glycosylated A chain (data not shown), excluding the possibility that the glycosylated toxin could be more firmly bound to the cells than the unglycosylated counterpart. The data indicate that, upon incubation with cells, ricin A chain containing exposed glycosylation sites becomes core glycosylated, presumably in the ER, and that maturation of the oligosaccharides does not occur.
To test to what extent sulfate-labeled ricin becomes glycosylated, we incubated cells with 35SO42− and ricin as in Fig. 3, and then washed the cells and incubated them further in medium without ricin and radioactive sulfate. The results in Fig. 4 show that although at least half of the toxin was in the unglycosylated form immediately after the change of medium (lane 1), most was glycosylated after 2.5 h (lane 2). After 5 h (lane 3), the total amount was somewhat reduced. Longer incubations resulted in toxin-induced cell death.
Figure 4.
Time course of glycosylation of sulfate-labeled ricin. Ricin A-sulf-2 reconstituted with ricin B chain was incubated with Vero cells for 4 h in the presence of 35SO42−, then the cells were washed and incubated further in medium without toxin and isotope. After the indicated periods of time, cells were harvested and analyzed by SDS/PAGE under nonreducing conditions.
Translocation of Ricin A Chain from the ER to the Cytosol.
To see if the labeled ricin A chain was released into the cytosol, it was necessary to open up the cells in such a way that the content in the Golgi apparatus and in the ER did not leak out into the cytosol fraction. To test this, we studied the leakage into the extraction buffer of cytosolic proteins as well as of proteins located inside organelles.
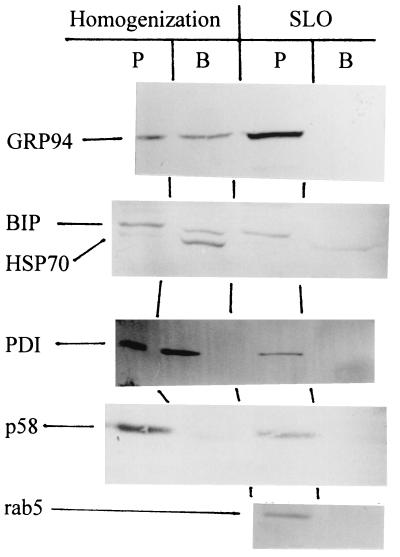
When the cells were disrupted mechanically and separated into a soluble and a pellet fraction by ultracentrifugation, 95% of the cytosolic enzyme lactate dehydrogenase was in the soluble fraction, indicating efficient homogenization. The two fractions were also submitted to SDS/PAGE, transferred to membranes that were probed with antibodies against the ER-located glucose-regulated proteins GRP94 (36) and GRP78 (or BIP; ref 37) and PDI (38). As shown in Fig. 5, these proteins were recovered partly from the pellet fractions and partly from the supernatant, indicating that a certain amount of organelle disruption had taken place during the homogenization. The intermediate compartment/Golgi-located protein, p58 (39), which is anchored to the membrane by a transmembrane stretch, was recovered mainly from the membrane fraction. The cytosol-located HSP70 (40) was recovered from the supernatant.
Figure 5.
Effect of cell permeabilization on release of resident organellar proteins. Vero cells were either homogenized or permeabilized with SLO as described in Materials and Methods. For homogenization, the cells were removed from the plastic by scraping and disrupted in PBS by passing them 10 times up and down through a 0.5-mm gauge syringe. The suspension was then centrifuged for 10 min in an Eppendorf centrifuge to remove nuclei and mitochondria. The postmitochondrial supernatant as well as the buffer fraction obtained after the SLO treatment were centrifuged at 128,000 × g for 30 min. In both cases, the pellet (P) was taken as the membrane fraction and the supernatant (B, buffer) as the cytosolic fraction. Both fractions were fractionated by SDS/PAGE, transferred to Immobilon membranes, and probed with antibodies against GRP94, BIP, HSP70, PDI, p58, and rab5.
To open the cells without disrupting intracellular organelles, we used SLO to selectively form pores in the plasma membrane (41, 42). SLO requires reducing agents to be active and to prevent reduction of intracellular toxin we used the membrane-impermeable MesNA (β-mercaptoethane sulfonate; ref. 43) as reducing agent. We bound SLO to cells at 4°C to ensure exclusive binding to the plasma membrane, then we washed the cells to remove free SLO, and finally we heated the cells briefly to 37°C to allow the surface-bound SLO to form pores in the plasma membrane. As shown in Fig. 5, under these conditions the tested protein markers were not released from the cells, indicating that the organelles remained intact. Also the cytoplasmic, but vesicle-associated, rab5 remained in the cellular pellet. On the other hand, the free cytosolic protein HSP70 was recovered in the buffer fraction, indicating that the cells were efficiently permeabilized. Also, 88% of the cytosolic lactate dehydrogenase was recovered in the buffer (data not shown).
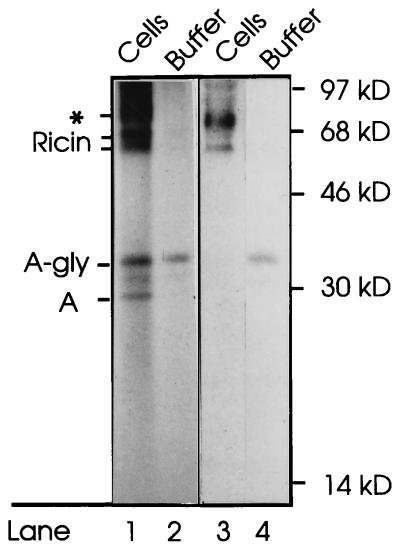
We then incubated cells for 4 h with 35SO4− and reconstituted toxin containing ricin A-sulf-2 and then treated the cells with N-ethylmaleimide to quench free SH-groups. Subsequently, the cells were washed with lactose to remove surface-bound ricin. After permeabilization with SLO, we collected the extracellular medium as well as the cells, which were subsequently lysed with Triton X-100 and centrifuged to remove the nuclei. Both supernatant fractions were treated with immobilized anti-ricin and analyzed by nonreducing SDS/PAGE. Whereas essentially all holotoxin remained in the cellular fraction (Fig. 6 (lanes 1 and 3), a considerable part of the free glycosylated A chain was found in the buffer (lanes 2 and 4). The amount present in the buffer fraction varied between experiments (compare the two experiments shown in lanes 1 and 2 and lanes 3 and 4). The results indicate that the glycosylated A chain had diffused out from the cytosol upon permeabilization of the cells. It is unlikely that the glycosylated A chain could be derived from labeled holotoxin that had been transported back to the surface membrane and subsequently reduced by MesNA, since it appears that only the nonglycosylated form of the toxin is exocytosed (Fig. 3a).
Figure 6.
Translocation of ricin A chain to the cytosol. Vero cells were incubated with Na235SO4 and ricin containing A-sulf-2 for 4 h. Then N-ethylmaleimide (10 mM) was added and the cells were further incubated for 10 min and then washed twice with PBS containing 0.1 M lactose. The cells were permeabilized with SLO. After collection of the buffer, the cells were lysed in Triton X-100 and centrifuged to remove the nuclei. Both supernatant fractions were treated with immobilized anti-ricin, and the adsorbed material was submitted to SDS/PAGE under nonreducing conditions. Lanes 1 and 2, and lanes 3 and 4 represent two different experiments. On the left, the positions of glycosylated and unglycosylated ricin holotoxin, and of glycosylated and unglycosylated free ricin A-sulf-2 are indicated. The band marked by an asterisk indicates the position of a contaminant, presumably a sticky sulfate-containing proteoglycan, that often comes down together with the immunoprecipitate. Molecular mass markers are indicated on the right.
DISCUSSION
The most important finding in the present work is that a mutant ricin A chain is translocated to the cytosol after first being transported to the ER. Glycosylation of the A chain does not appear to interfere with the translocation. This may not be too surprising since natural ricin A chain contains two asparagine-linked oligosaccharide chains (32), and toxin containing recombinant, unglycosylated A chain is not more toxic than natural ricin.
A central question is how ricin having reached the Golgi apparatus is transported retrograde to the ER. Since the toxin does not contain a KDEL-like retrieval sequence as some other toxins do (16), it is likely that either molecules by which ricin is bound and endocytosed contain a sequence directing it to the ER or, alternatively, that the toxin becomes detached from its primary binding site in the trans-Golgi network and subsequently bound by its B chain to molecules that shuttle between Golgi and ER (44, 45). In fact, there is evidence that the galactose-binding site of ricin is required even when the toxin is bound to the cells by an alternative binding mechanism (46, 47).
Only about half of ricin A-sulf-2 was glycosylated at any time in experiments where the cells were incubated continuously with ricin and radioactive sulfate. In chase experiments, however, it appeared that essentially all 35SO4−-labeled ricin associated with the cells becomes eventually glycosylated. The delay in glycosylation probably reflects the time required for transport from the Golgi apparatus to the ER, but it is also possible that the protein is not glycosylated immediately upon reaching the ER. The ER seems the most likely compartment for translocation of the A chain to the cytosol since it is known that transport of many large molecules, including peptides, occurs in both directions across the ER membrane (48). There is also evidence that proteins can be translocated from the ER to the cytosol. Human cytomegalovirus down-regulates glycosylated major histocompatibility complex class I molecules by proteolysis involving proteasomes. Proteasomes are found in the cytosol and in the nucleus. A resident type-1 transmembrane protein, the product of the US11 gene of the virus, is required for this process and could represent a transport protein (49). In yeast, glycosylated carboxypeptidase Y, which is present in the lumen of the ER, appears to be transported back into the cytosol to be degraded by an ubiquitin- and proteasome-dependent mechanism (50). It is possible that a transmembrane ER protein, Der1, is involved in the transport (51). Also a mutant variant of human α-1-proteinase inhibitor expressed in yeast appears to be transported out of the ER to be degraded by proteasomes (52). Possibly, ricin A chain takes advantage of a preexisting transport mechanism to escape into the cytosol. Although this mechanism may have been developed for degradation by proteasomes of proteins that are misfolded in the ER, the translocated ricin A chain may have acquired the ability to avoid the proteasomes for sufficient time to carry out its enzymatic effect on the ribosomes.
Only part of the labeled and glycosylated ricin is reduced and translocated to the cytosol. Similar findings have earlier been made with diphtheria toxin (53). Also in that case the fraction that is translocated varies between experiments (53, 54). In cases like ricin, where a preexisting translocation mechanism may be used, it appears likely that the translocation is incomplete. It is not clear if the reduction of the interchain disulfide in ricin occurs concomitantly with translocation as in the case of diphtheria toxin (55), or if the reduction occurs in the ER independently of translocation. Possibly, only A chains that are already released from the B chain can engage the translocation apparatus. The finding that, in some experiments, less that half of the free A chain in the cells was released into the buffer under conditions where the major part of the cytosolic proteins HSP70 and lactate dehydrogenase was released suggests that the remaining free A chain is contained inside the ER.
The possibility that reduced A chain could leak out from damaged ER is unlikely since the proteins GRP94, BIP, and PDI, which are located free in the ER lumen, were not released into the cytoplasmic fraction upon permeabilization of the cells with SLO. On the other hand, when the cells were disrupted mechanically, about the half of each of these proteins was recovered from the cytosolic fraction. Although part of these proteins, which all have a C-terminal KDEL ER-retrieval sequence, may be bound to the KDEL receptor (56), this binding would be of low affinity at pH 7.5 in the permeabilization buffer. Also the cytoplasmic, but vesicle-associated, rab5 protein was not released by the SLO treatment. p58, which is located in the intermediary compartment between the ER and Golgi, was not released to a large extent even after homogenization. This is consistent with the fact that p58 is a transmembrane protein that may remain attached even to disrupted fragments of the organelle.
If a preexisting mechanism is used for translocation of ricin A chain, the B chain may not be involved in the process. This would be different from diphtheria toxin where the B fragment appears to form a major part of the translocation apparatus (57). Diphtheria toxin is translocated from endosomes, and the translocation occurs as soon as sufficient acidification of the vesicles has taken place to allow insertion of the toxin B fragment into the membrane (17). If ricin and a number of other toxins must be transported retrograde to the ER because they lack an intrinsic translocation apparatus, the observation that the B chain of ricin and other toxins can be replaced by antibodies against cell surface molecules to form active immunotoxins would be more understandable. In this case, the antibody would only have to ensure binding to a molecule that is endocytosed and transported retrograde to the ER. The large difference in immunotoxin activity could reflect different efficiency with which the binding molecule is transported retrograde in the cells.
Acknowledgments
We are indebted to Dr. M. Monsigny for suggesting the addition of terminal glycosylation sites onto the A chain and to Dr. H. Stenmark for critical reading of the manuscript. This work was supported by The Norwegian Cancer Society, Novo Nordisk Foundation, The Norwegian Research Council for Science and Humanities, Blix Legat, Rachel and Otto Kr. Bruun’s Legat, and The Jahre Foundation. A.R. and P.Ø.F. are Postdoctoral Fellows of The Norwegian Cancer Society.
ABBREVIATIONS
- ER
endoplasmic reticulum
- SLO
streptolysin O
- PDI
protein disulfate isomerase
- MBP
maltose-binding protein
- endo H
endoglycosidase H
- PNGase F
endoglycosidase F
- HSP70
heat shock protein 70
- GRP94
glucose regulated protein 94
References
- 1.Olsnes S, Sandvig K, Petersen O W, van Deurs B. Immunol Today. 1989;10:291–295. [PubMed] [Google Scholar]
- 2.Vitetta E S, Thorpe P E. Semin Cell Biol. 1991;2:47–58. [PubMed] [Google Scholar]
- 3.Thrush G R, Lark L R, Clinchy B C, Vitetta E S. Annu Rev Immunol. 1996;14:49–71. doi: 10.1146/annurev.immunol.14.1.49. [DOI] [PubMed] [Google Scholar]
- 4.Olsnes S, Pihl A. Biochemistry. 1973;12:3121–3126. doi: 10.1021/bi00740a028. [DOI] [PubMed] [Google Scholar]
- 5.Endo Y, Mitsui K, Motizuki M, Tsurugi K. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 6.Endo Y, Tsurugi K. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 7.Olsnes S, Refsnes K, Pihl A. Nature (London) 1974;249:627–631. doi: 10.1038/249627a0. [DOI] [PubMed] [Google Scholar]
- 8.Sandvig K, Olsnes S, Pihl A. J Biol Chem. 1976;251:3977–3984. [PubMed] [Google Scholar]
- 9.van Deurs B, Tønnessen T I, Petersen O W, Sandvig K, Olsnes S. J Cell Biol. 1986;102:37–47. doi: 10.1083/jcb.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wales R, Chaddock J A, Roberts L M, Lord J M. Exp Cell Res. 1992;203:1–4. doi: 10.1016/0014-4827(92)90032-4. [DOI] [PubMed] [Google Scholar]
- 11.Wales R, Roberts L M, Lord J M. J Biol Chem. 1993;268:23986–23990. [PubMed] [Google Scholar]
- 12.Tagge E, Chandler J, Tang B L, Hong W J, Willingham M C, Frankel A. J Histochem Cytochem. 1996;44:159–165. doi: 10.1177/44.2.8609372. [DOI] [PubMed] [Google Scholar]
- 13.Reisbig R, Olsnes S, Eiklid K. J Biol Chem. 1981;256:8739–8744. [PubMed] [Google Scholar]
- 14.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. Eur J Biochem. 1988;171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- 15.Sandvig K, Garred Ø, Prydz K, Kozlov J V, Hansen S H, van Deurs B. Nature (London) 1992;358:510–512. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhary V K, Jinno Y, FitzGerald D, Pastan I. Proc Natl Acad Sci USA. 1990;87:308–312. doi: 10.1073/pnas.87.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsnes S, Sandvig K. In: Immunotoxins. Frankel A E, editor. Boston: Kluwer; 1988. pp. 39–73. [Google Scholar]
- 18.Pelham H R B, Roberts L, Lord J M. Trends Cell Biol. 1992;2:183–185. doi: 10.1016/0962-8924(92)90230-k. [DOI] [PubMed] [Google Scholar]
- 19.van Deurs B, Sandvig K, Petersen O W, Olsnes S, Simons K, Griffiths G. J Cell Biol. 1988;106:253–267. doi: 10.1083/jcb.106.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitinger B, Brown J L, Spiess M. J Biol Chem. 1994;269:8115–8121. [PubMed] [Google Scholar]
- 21.Sundan A, Evensen G, Hornes E, Mathiesen A. Nucleic Acids Res. 1989;17:1717–1732. doi: 10.1093/nar/17.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Elbein A D. Methods Enzymol. 1983;98:135–154. doi: 10.1016/0076-6879(83)98144-2. [DOI] [PubMed] [Google Scholar]
- 24.Kasinathan C, Sundaram P, Slomiany B L, Slomiany A. Biochemistry. 1993;32:1194–1198. doi: 10.1021/bi00055a026. [DOI] [PubMed] [Google Scholar]
- 25.Sandvig K, Tønnessen T I, Olsnes S. Cancer Res. 1986;46:6418–6422. [PubMed] [Google Scholar]
- 26.Halstead J, Kemp K, Ignotz R A. J Biol Chem. 1995;270:13600–13603. doi: 10.1074/jbc.270.23.13600. [DOI] [PubMed] [Google Scholar]
- 27.Sandvig K, Olsnes S. J Biol Chem. 1982;257:7504–7513. [PubMed] [Google Scholar]
- 28.Veit B, Yucel J K, Malhotra V. J Cell Biol. 1993;122:1197–1206. doi: 10.1083/jcb.122.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acharya U, McCaffery J M, Jacobs R, Malhotra V. J Cell Biol. 1995;129:577–589. doi: 10.1083/jcb.129.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandvig K, Prydz K, Hansen S H, van Deurs B. J Cell Biol. 1991;115:971–981. doi: 10.1083/jcb.115.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nambiar M P, Wu H C. Exp Cell Res. 1995;219:671–678. doi: 10.1006/excr.1995.1278. [DOI] [PubMed] [Google Scholar]
- 32.Olsnes S, Pihl A. Mol Action Toxins Viruses. 1982;3:51–105. [Google Scholar]
- 33.Rosa P, Mantovani S, Rosboch R, Huttner W B. J Biol Chem. 1992;267:12227–12232. [PubMed] [Google Scholar]
- 34.Cole N, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balch W E, Fries E, Dunphy W H, Urbani L J, Rothman J E. Methods Enzymol. 1983;98:37–47. doi: 10.1016/0076-6879(83)98137-5. [DOI] [PubMed] [Google Scholar]
- 36.Sorger P K, Pelham H R. J Mol Biol. 1987;194:341–344. doi: 10.1016/0022-2836(87)90380-9. [DOI] [PubMed] [Google Scholar]
- 37.Munro S, Pelham H R. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 38.Freedman R B, Hawkins H C, Murant S J, Reid L. Biochem Soc Trans. 1988;16:96–99. doi: 10.1042/bst0160096. [DOI] [PubMed] [Google Scholar]
- 39.Saraste J, Svensson K. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- 40.Pelham H. Nature (London) 1988;332:776–777. doi: 10.1038/332776a0. [DOI] [PubMed] [Google Scholar]
- 41.Bhakdi S, Bayley H, Valeva A, Valev I, Walker B, Weller U, Kehoe M, Palmer M. Arch Microbiol. 1996;165:73–79. doi: 10.1007/s002030050300. [DOI] [PubMed] [Google Scholar]
- 42.Bhakdi S, Weller U, Walev I, Martin E, Jonas D, Palmer M. Med Microbiol Immunol (Berl) 1993;182:167–175. doi: 10.1007/BF00219946. [DOI] [PubMed] [Google Scholar]
- 43.Carter L, Redelmeier T, Woollenweber L, Schmid S. J Cell Biol. 1993;120:37–45. doi: 10.1083/jcb.120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson J C, Dascher C, Roberts L M, Lord J M, Balch W E. J Biol Chem. 1995;270:20078–20083. doi: 10.1074/jbc.270.34.20078. [DOI] [PubMed] [Google Scholar]
- 45.Miesenbock G, Rothman J E. J Cell Biol. 1995;129:309–319. doi: 10.1083/jcb.129.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Youle R J, Murray G J, Neville D M., Jr Cell. 1981;23:551–559. doi: 10.1016/0092-8674(81)90151-3. [DOI] [PubMed] [Google Scholar]
- 47.Newton D L, Wales R, Richardson P T, Walbridge S, Saxena S K, Ackerman E J, Roberts L M, Lord J M, Youle R J. J Biol Chem. 1992;267:11917–11922. [PubMed] [Google Scholar]
- 48.Römisch K. Trends Cell Biol. 1994;4:311–314. doi: 10.1016/0962-8924(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 49.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 50.Hiller M, Finger A, Schweiger M, Wolf D. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 51.Knop M, Finger A, Braun T, Hellmuth K, Wolf D H. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 52.Werner E D, Brodsky J L, McCracken A A. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moskaug J O, Sandvig K, Olsnes S. J Biol Chem. 1988;263:2518–2525. [PubMed] [Google Scholar]
- 54.Falnes P Ø, Choe S, Madshus I H, Wilson B A, Olsnes S. J Biol Chem. 1994;269:8402–8407. [PubMed] [Google Scholar]
- 55.Falnes P Ø, Olsnes S. J Biol Chem. 1995;270:20787–20793. doi: 10.1074/jbc.270.35.20787. [DOI] [PubMed] [Google Scholar]
- 56.Scheel A, Pelham R. Biochemistry. 1996;35:10203–10209. doi: 10.1021/bi960807x. [DOI] [PubMed] [Google Scholar]
- 57.O’Keefe D O, Cabiaux V, Choe S, Eisenberg D, Collier R J. Proc Natl Acad Sci USA. 1992;89:6202–6206. doi: 10.1073/pnas.89.13.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]